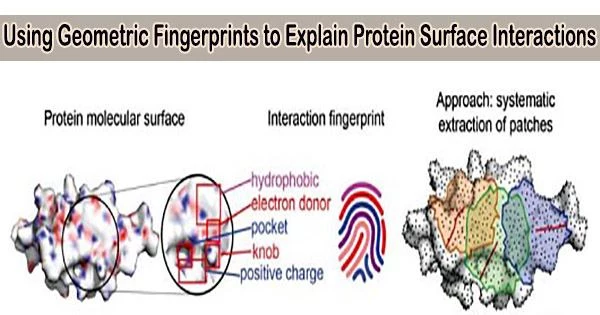
Researchers from the Swiss Institute of Bioinformatics in Lausanne, Switzerland, described geometric and chemical properties crucial to protein-protein interactions using a geometric deep-learning technology that creates “fingerprints” of protein surfaces.
In their paper, “De novo design of protein interactions with learned surface fingerprints,” published in Nature, the team report that their hypothesized “fingerprints” led to the capture of essential aspects of molecular recognition and novel protein interactions. The team’s findings are compiled in a Research Briefing that is printed in the same journal issue.
Proteins are the biological system’s cogs, gears, springs, and valves that make organic life possible. It is where most diseases connect with biological systems, where cells function, and how medications engage with biological systems. Science could treat the majority of diseases if it had a commanding understanding of this equipment.
With one small but important caveat, the binding sites, machine learning, proteomics, genomic sequencing, and molecular biology have significantly accelerated protein research over the past ten years.
Today, we can predict nearly all functional protein structures, engineer novel structures in any configuration, and synthetically produce any protein we can think of.
Two distinct physical “lock-and-key” interactions based on surface chemistry and structure govern how proteins interact. Both an upper, buried site and an outer rim site exist. The protein’s function is carried out by the buried site, but in order to access it, the initial outer rim signal is needed to crack open the structure.
The affinities of proteins have remained elusive despite the advances made in proteomics over the past ten years. This is partially due to the fact that proteins are picky and pH-dependent, with variable rim surface chemistry and condition and site-specific binding sites.
In the current study, the scientists focused solely on the surface affinity while pursuing structural affinity between proteins. The team concentrated the machine learning on the surface interactions of proteins and the geometric and chemical patterns that determine the best chance for two molecules to interact, then designed the appropriate keys, putting aside information about the overall structure, function, and similar protein interactions to their targets.
The team was able to quickly and accurately locate complimentary surface fragments that can engage a particular target among 402 million possible surfaces by computing fingerprints from protein molecular surfaces.
Several de novo protein binders were computationally designed to engage four protein targets: SARS-CoV-2 spike, PD-1, PD-L1 and CTLA-4. While some ideas were created entirely in the digital realm, others were optimized through experimentation.
As a result of the machine learning-based binders successfully engaging their targets, the results were extremely accurate affinity predictions.
The authors state that their framework could “…open possibilities in other important biotechnological fields such as drug design, biosensing or biomaterials in addition to providing a means to study interaction networks in biological processes at the systems levels.”