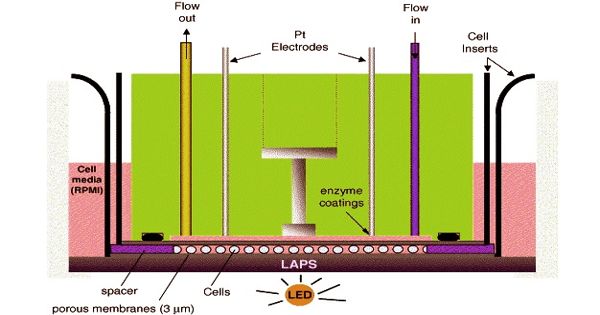
Microphysiometry has proved to be a useful tool for monitoring the energy metabolism of living cells and their interactions with other cells. It is the in vitro measurement of the functions and activities of life or of living matter (as organs, tissues, or cells) and of the physical and chemical phenomena involved on a very small (micrometer) scale. Time-resolved monitoring of metabolic activities in vitro (microphysiometry) is necessary to study the dynamic regulations of core biochemical pathways in cellular disease models. The term microphysiometry emerged in the scientific literature at the end of the 1980s.
The primary parameters assessed in microphysiometry comprise pH and the concentration of dissolved oxygen, glucose, and lactic acid, with an emphasis on the first two. The technique has mainly been used for monitoring two-dimensional (2D) monolayers of cells. The dynamic activity of the cellular core metabolism is a key functional parameter for the assessment of living cells. The capability of continuously monitoring cells and tissues in real-time for hours or days supports the value of a sensor-based microphysiometric approach.
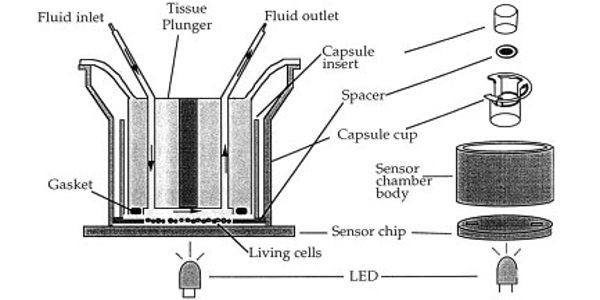
Microphysiometry is a non-invasive, physiological method where measurement of metabolic activity can be made on living human tumor cells. Indirect measurement of extracellular acidification is measured over a pH-sensitive silicon membrane. Measuring these parameters experimentally in combination with a fluidic system for cell culture maintenance and a defined application of drugs or toxins provides the quantitative output parameters extracellular acidification rates (EAR), oxygen consumption rates (OUR), and rates of glucose consumption or lactate release to characterize the metabolic situation. However, applying this technique for automated label-free and real-time measurements within cell-laden three-dimensional (3D) cell culture constructs remains a challenge.
Microphysiometry is a powerful technique to study metabolic parameters and detect changes to external stimuli. Due to the label-free nature of sensor-based measurements, dynamic monitoring of cells or tissues for several days or even longer is feasible. Metabolic adaptation to stress is a crucial yet poorly understood phenomenon, particularly in the central nervous system (CNS). On an extended timescale, a dynamic analysis of a cell’s metabolic response to an experimental treatment can distinguish acute effects (e.g., one hour after treatment), early effects (e.g., at 24 hours), and delayed, chronic responses (e.g., at 96 hours). Most of the investigated drugs showed some initial stimulation of acidification rate during the experiments. The ability to identify essential metabolic events which predict neuronal fate in response to injury is critical to developing predictive markers of the outcome, for interpreting CNS spectroscopic imaging, and for providing a richer understanding of the relevance of clinical indices of stress which are routinely collected.
Information Source: