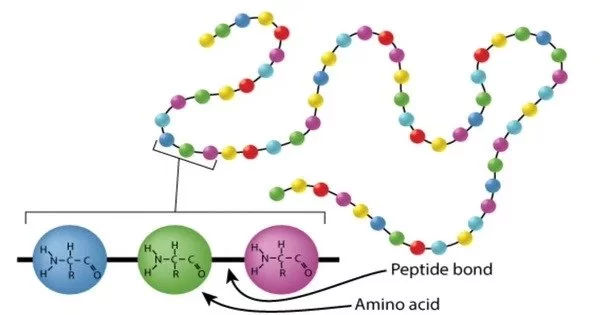
Protein sequencing is the technique of determining the amino acid sequence of a protein or peptide in its entirety or in part. It is the process of determining a protein’s amino acid sequence. This could be used to discover or characterize the protein’s post-translational modifications. Partial sequencing of a protein typically offers enough information (one or more sequence tags) to identify it using databases of protein sequences produced via conceptual translation of genes. The amino acid sequence of a protein is critical for understanding its structure, function, and interactions with other molecules in biological processes.
Mass spectrometry and Edman degradation utilizing a protein sequenator (sequencer) are the two basic direct methods of protein sequencing. Although mass spectrometry is now the most extensively used approach for protein sequencing and identification, Edman degradation is still a useful tool for identifying a protein’s N-terminus.
Here’s an overview of protein sequencing techniques and methods:
- Edman Degradation: One of the first and most extensively used approaches for protein sequencing is the Edman degradation method. It entails deleting an N-terminal amino acid from a protein one at a time. This method is repeated until the complete protein sequence is known. It is, however, time-consuming and may not be appropriate for big proteins.
- Mass Spectrometry (MS): For protein sequencing and identification, mass spectrometry is a strong approach. A protein is enzymatically digested into smaller peptides, and the weights of these peptides are determined using a mass spectrometer in this procedure. Researchers can identify the protein and, in some cases, ascertain its sequence by evaluating the mass-to-charge ratios of these peptides and comparing them to a protein sequence database.
- Next-Generation Sequencing (NGS): NGS technologies, such as Illumina sequencing, are primarily used for DNA and RNA sequencing. However, they can also be adapted for protein sequencing by first converting proteins into complementary DNA (cDNA) through reverse transcription. This approach is less common than other methods but has been used for some protein sequencing applications.
- Chemical Cleavage Methods: Various chemical methods can be used to cleave proteins at specific amino acid residues. For example, cyanogen bromide (CNBr) can cleave proteins at methionine residues, and trypsin can cleave proteins at lysine and arginine residues. By using a combination of chemical cleavage methods and mass spectrometry, researchers can deduce the protein sequence.
- NMR Spectroscopy: NMR spectroscopy can be used to determine the three-dimensional structure of proteins, which can reveal information about the amino acid sequence indirectly by studying the connectivity of atoms in the protein.
- X-ray Crystallography: Another approach that can reveal detailed information on a protein’s three-dimensional structure is X-ray crystallography. While it does not directly determine the amino acid sequence, it can be used to validate and modify protein sequences in conjunction with other approaches.
Each protein sequencing approach has advantages and disadvantages, and the technology used is determined by criteria such as protein size, available resources, and study objectives. Because of its speed and sensitivity, mass spectrometry has grown in popularity and is now commonly employed in modern proteomics research.