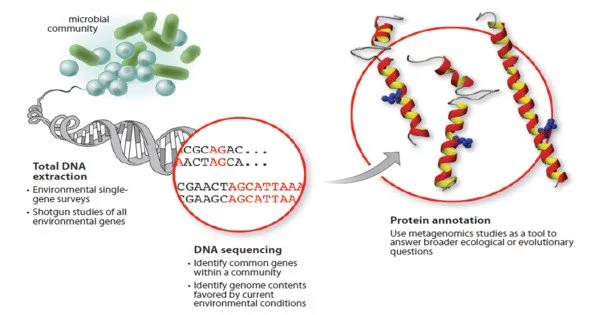
The study of genetic material collected directly from environmental or clinical samples using a technology known as sequencing is known as metagenomics. It is a branch of molecular biology that focuses on the examination of genetic material extracted directly from environmental samples. Environmental genomics, ecogenomics, community genomics, and microbiomics are all terms used to describe the broad area. Unlike classical genomics, which focuses on individual species’ genomes, metagenomics involves the study of genetic material collected from a community of microbes, such as those found in soil, water, or the human gut.
While classical microbiology and microbial genome sequencing and genomics rely on clonal cultures, early environmental gene sequencing cloned particular genes (typically the 16S rRNA gene) to generate a diversity profile in a natural sample. Such research demonstrated that cultivation-based methods have missed the vast majority of microbial biodiversity.
Metagenomics offers a strong approach of comprehending the microbial world that may transform understanding of biology due to its capacity to disclose previously unknown diversity of tiny life. As the cost of DNA sequencing continues to decline, metagenomics permits microbial ecology to be studied at a far larger scale and in greater detail than before. Recent studies use either “shotgun” or PCR directed sequencing to get largely unbiased samples of all genes from all the members of the sampled communities.
Metagenomics’ major purpose is to study the structure and function of microbial communities within a specific environment. Researchers can use this method to investigate the collective genomes of several organisms at the same time, revealing insights into the variety and dynamics of microbial populations, their interactions, and their roles in various ecological processes.
Metagenomic studies typically involve the following steps:
- Sampling: Collecting samples from various environmental sources, such as soil, water, or the human body.
- DNA Extraction: Isolating and extracting genetic material, including DNA, from the collected samples.
- Sequencing: Performing high-throughput DNA sequencing to determine the genetic composition of the sample. This step produces a vast amount of sequence data.
- Bioinformatics Analysis: Using various computational tools and bioinformatics techniques, analyze the produced sequence data to identify and characterize the microbial species present in the sample, as well as their functional capabilities and relationships.
Metagenomics offers a wide range of applications, including environmental microbiology, biotechnology, and human health. It has expanded our understanding of microbial diversity, evolution, and the functions of microbes in ecological processes, nutrient cycling, and disease development dramatically. Metagenomics has also resulted in the discovery of new genes, enzymes, and natural products with potential applications in medicine, agriculture, and bioremediation.