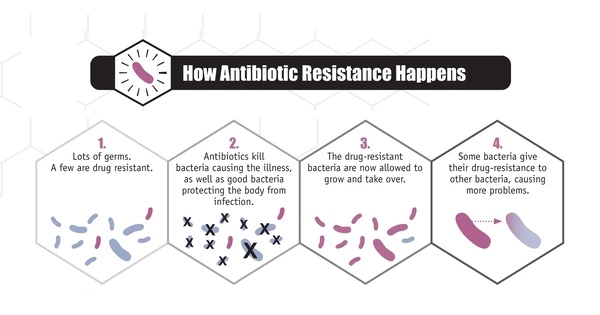
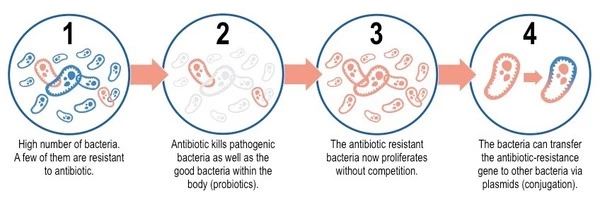
Bacteria reproduce quickly, and during replication, errors in their genetic material can occur, resulting in mutations. These mutations can occasionally provide bacteria with antibiotic resistance. Bacteria may become resistant if a mutation changes the target of an antibiotic or the pathway involved in its uptake or metabolism.
According to new research from the Quadram Institute and the University of East Anglia, bacteria can rapidly evolve antibiotic resistance by adapting special pumps to flush them out of their cells.
Antimicrobial resistance is a growing issue with global implications. The emergence of resistant “superbugs” jeopardizes our ability to treat and prevent the spread of infections caused by microorganisms using antimicrobials such as antibiotics. The findings are hoped to improve how antibiotics are used in order to help prevent the spread of antimicrobial resistance.
“Knowing the details of the mechanisms bacteria develop to become resistant is a key step to understanding antimicrobial resistance,” said Prof Mark Webber of UEA’s Norwich Medical School and the Quadram Institute. We hope that this type of research into when and how resistance emerges will help us use antibiotics more effectively and reduce resistance selection.”
This work simulates what happens in the real world where bacteria are constantly exposed to varying concentrations of antimicrobials. Understanding how resistant strains emerge and which drugs they will not respond to can aid in the development of diagnostics and treatment strategies.
Dr. Eleftheria Trampari
The researchers investigated how antimicrobial exposure leads to the emergence of resistance. In general, superbugs’ antibiotic defenses involve inactivating or evading drugs, preventing them from entering their cells, or removing them from their cells before they can have any effect. However, the specifics of how they will accomplish this are still being worked out.
Dr Eleftheria Trampari of QI, Prof Webber, and colleagues recreated the evolutionary stresses that lead to antimicrobial resistance by exposing Salmonella bacteria to two different antibiotics in this new study.
The bacteria were allowed to grow and reproduce in two different states that mimic how they live in the environment. Some were planktonic — floating in a liquid broth — but others were in biofilms. Bacteria form biofilms on surfaces, as a way of protecting themselves against stresses and most bacteria in the real world exist in a biofilm.
Hundreds of generations of bacteria were grown and exposed to the antibiotics, and in this evolution simulation, survival of the fittest selected those bacteria best adapted to cope with the presence of the antibiotics.
The researchers sequenced the genomes of the resistant bacteria to determine which genes had changed compared to their non-resistant ancestors in order to determine how these ‘winners’ had become resistant.
Both antibiotics selected different mutations in a molecular pump that Salmonella uses to remove toxic compounds from its cells. They discovered, along with colleagues from the Universities of Essex and Cagliari, that these two different changes altered how the pump worked in completely different ways. One made it easier for drugs to be caught by the pumps, while the other made it easier for drugs to pass through the pump.
A search of Salmonella genome databases revealed that one of these mutations has appeared multiple times in the real world, in Salmonella from patients, livestock, and food in the UK, US, and EU, dating back to 2003.
The findings support these pumps’ primary role as the first line of defense against antimicrobials.
“This work simulates what happens in the real world where bacteria are constantly exposed to varying concentrations of antimicrobials,” said Quadram Institute’s Dr Eleftheria Trampari, the study’s first author. “Understanding how resistant strains emerge and which drugs they will not respond to can aid in the development of diagnostics and treatment strategies.”