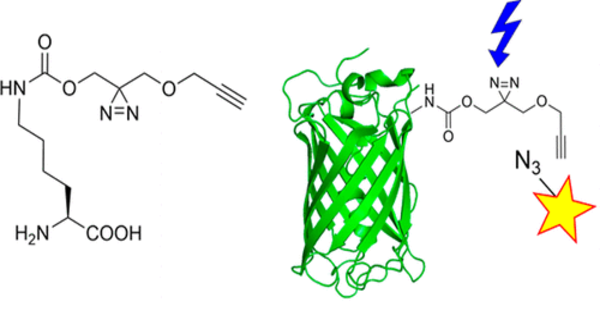
Unnatural amino acids are chemically altered versions of the 20 amino acids found in nature. They may have properties or functions that are not found in natural amino acids. Researchers can modify the bacteria’s metabolism to produce these new building blocks by introducing genes encoding enzymes capable of synthesizing these unnatural amino acids into the genetic code.
Researchers engineered bacteria to produce an amino acid containing a rare functional group that has previously been linked to immune system regulation. The researchers also taught a single bacterial strain to produce the amino acid and insert it into specific locations within target proteins. These findings lay the groundwork for the future development of novel vaccines and immunotherapies.
Amino acids are the basic building blocks of proteins and are required for biological systems to function properly. There are 20 standard amino acids found in all living systems that make up proteins, as well as over 500 different types of other amino acids found in nature and a large number of man-made amino acids. Some of these alternative amino acids may aid in the development of new pharmaceuticals and therapeutics.
Now, University of Delaware researchers in the lab of Aditya Kunjapur, assistant professor in the College of Engineering’s Department of Chemical and Biomolecular Engineering, have engineered bacteria to synthesize an amino acid that contains a rare functional group that others have shown to have implications in the regulation of our immune system. The researchers also taught a single bacterial strain to create the amino acid and place it at specific sites within target proteins. These findings, published in Nature Chemical Biology, provide a foundation for developing unique vaccines and immunotherapies in the future.
Bacteria are potentially useful drug delivery vehicles. We believe we have developed a tool that can take advantage of bacteria’s ability to produce target antigens within the body while also utilizing nitration’s ability to shine a light on those antigens.
Aditya Kunjapur
The Kunjapur Lab creates microorganisms that can synthesize various types of compounds and molecules, particularly those with functional groups or properties that are not well represented in nature, using tools from synthetic biology and genetic engineering.
The researchers focused on para-nitro-L-phenylalanine (pN-Phe), a non-standard amino acid that has neither been observed in nature nor is one of the twenty standard amino acids. Other research groups have used pN-Phe to help the immune system mount a response to proteins that it normally does not respond to.
“The nitro chemical functional group has valuable properties and has been underexplored by folks who are trying to rewire metabolism,” Kunjapur said. “pN-Phe also has a nice history in the literature — it can be added onto a protein from a mouse, delivered back to mice, and the immune system will no longer tolerate the original version of that protein. That ability has promise for the treatment or prevention of diseases that are caused by rogue proteins that the immune system struggles to lock onto.”
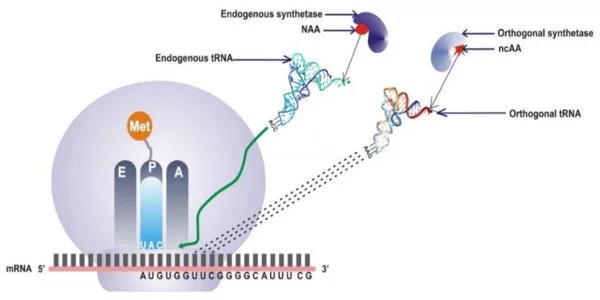
This paper describes the first demonstration of pN-Phe synthesis in Escherichia coli and is one of only a few that feature an organism designed to autonomously create and harness an expanded genetic code. The researchers were able to increase the “alphabet” of available amino acids encoded by DNA by using genetic code expansion methods. The researchers were able to create a system that produces nitrated proteins on its own by combining metabolic engineering techniques with genetic code expansion.
“Because of the nitro functional group chemistry, the amino acid that we chose as our target for this project was unconventional,” Kunjapur explained. “Many scientists in our field may not have expected that it could be made using biosynthesis.”
The next step for this research is to optimize their methods to synthesize higher amounts of nitrated proteins and expand this work into other microorganisms. The long-term goal is to further refine this platform for applications related to vaccines or immunotherapies, efforts that are supported by Kunjapur’s 2021 AIChE Langer Prize and the 2022 National Institutes of Health Director’s New Innovator Award. To further support this long-term goal, Kunjapur and Neil Butler, doctoral candidate and first author on this paper, co-founded Nitro Biosciences.
“I think the implications are interesting in that you can take a bacterium’s central metabolism, its ability to produce different compounds, and expand its chemical repertoire with a few modifications,” Butler said. “The nitro functionality is uncommon in biology and not found in the standard 20 amino acids, but we demonstrated that bacterial metabolism is malleable enough to be rewired to create and integrate this functionality.”
“Bacteria are potentially useful drug delivery vehicles,” Kunjapur added. We believe we have developed a tool that can take advantage of bacteria’s ability to produce target antigens within the body while also utilizing nitration’s ability to shine a light on those antigens.”