A proteome is the entire set of proteins that an organism expresses. The term can also refer to the collection of proteins produced at a specific time in a specific cell or tissue type. The proteome is an expression of the genome of an organism. In contrast to the genome, which is stable, the proteome changes in response to a variety of factors, including the organism’s developmental stage and both internal and external conditions.
Proteomics based on mass spectrometry is a big-data science of proteins that allows for the simultaneous monitoring of the abundance of thousands of proteins in a sample. As a result, it is an excellent readout for determining which proteins are targeted by any small molecule.
Histone deacetylase (HDAC) inhibitors are a type of oncology drug. Scientists from the Technical University of Munich (TUM), Cornell University in Ithaca (USA), the German Cancer Research Center (DKFZ) in Heidelberg, and Martin Luther University in Halle-Wittenberg have now studied the effects of some HDAC drugs in greater depth. The researchers investigated whether the epidrugs affect proteins other than the HDACs that they are intended to inhibit.
We’re excited because we’ve discovered a new player in this field of biology that includes exosomes, which play important roles in neurology, immunology, and oncology. We are now designing molecules that only target MBLAC2 in order to investigate this obscure protein in a variety of model systems.
Dr. Guillaume Médard
“To do so, target deconvolution by chemical proteomics is the method of choice. Hence, we first made new chemical tools — the so called affinity matrices — that would allow us to systematically profile the HDACs,” explains Dr. Guillaume Médard, group leader for chemical proteomics at the TUM chair of Proteomics and Bioanalytics led by Prof. Bernhard Küster.
Profiling HDAC drugs by chemical proteomics
“I profiled 53 drugs, and the majority of them, but not all, hit their intended HDAC target,” said Severin Lechner, a doctoral student at TUM’s School of Life Sciences. “However, there were some surprises. The drugs used in hundreds of scientific studies were not as selective as previously thought. Many of the targets had previously unknown targets.
These findings demonstrate the power of proteomic approaches, which can investigate the binding of thousands of proteins at once. Finally, the team discovered several molecules with high selectivity, making them ideal inhibitors for future scientific studies.
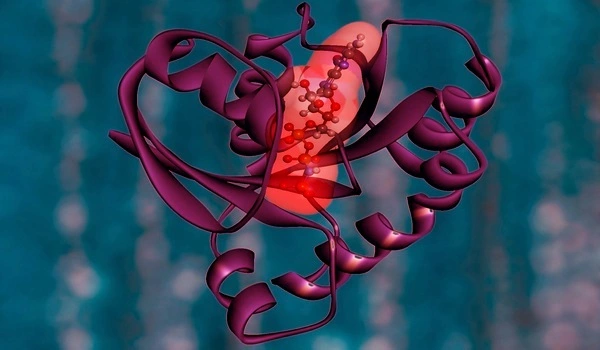
The target landscape of HDAC drugs
“The most surprising finding was that MBLAC2 is off-target for half of the molecules profiled,” Lechner continues. This protein is not well understood. At the same time, Prof. Maurine Linder’s team at Cornell was researching it. The two groups worked together to confirm that the protein is indeed hampered in its function in the presence of the drugs.
Working with Prof. Michael Pfaffl’s group at TUM, Lechner investigated the previously unexplained phenotypic effects of some drugs and demonstrated that MBLAC2 inhibition or knock-down causes an accumulation of extracellular vesicles in the extracellular space. Extracellular vesicles are small membrane-bound particles that are secreted by cells and transported throughout the body to transport biomolecules and information between cells and tissues.
Fundamental research to make tomorrow’s epidrugs
“We’re excited because we’ve discovered a new player in this field of biology that includes exosomes, which play important roles in neurology, immunology, and oncology,” Médard says. “We are now designing molecules that only target MBLAC2 in order to investigate this obscure protein in a variety of model systems.”
This research will be useful for those who want to use HDAC inhibitors to investigate biology or for therapeutic purposes. It aids in the selection of the appropriate chemical tool. It’s also a useful set of data for medicinal chemists, who need to understand how chemical structures relate to potency and selectivity in order to develop tomorrow’s epidrugs.