The manifestation of genetic features and their relationships are frequently influenced by biological processes, resource availability, and evolutionary processes. In fact, due to various ecological circumstances, a number of plant features are frequently connected. Height of plants and seed size are two typical examples of such connected features.
This correlation is broadly associated with the allometric scaling (the relative growth rates of different body parts versus the total body size of an organism) following a positive correlation.
A number of conceptual frameworks exist to explain the underlying mechanisms of allometric scaling in plants, one of which postulates that domestication and crop breeding in the modern age can change how plant height and seed weight evolve and are expressed.
The actual effects of intense domestication and breeding, as well as the genetic mechanisms determining size scaling and trait correlation in plants, have not yet been fully investigated.
The underlying genetic mechanisms of trait correlations and size scaling can be decoded using high-throughput genotyping and extensive phenotyping.
As a result, current research by Australian scientists has pinpointed the potential genetic components underpinning the association between plant height and seed weight scaling in barley harvests.
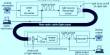
The study measured the variations in plant height and seed weight using a genome-wide single-nucleotide polymorphism (SNP) profile for a varied group of barley samples.
Our findings provide a better understanding of the genetic factors underlying size scaling, which, in turn, could expand the scope for exploring other mechanisms underlying the allometric scaling in plants. While our study examined size scaling in a single crop species, it points towards the possible existence of similar genomic mechanisms for allometric scaling across other plant species.
Professor Chengdao Li
Explaining the rationale for their study, Prof. Chengdao Li, the corresponding author of the study, from Murdoch University says, “Genome-wide association studies (GWASs) have shown that SNPs can help identify genetic variants affecting two or more traits. Moreover, once the physical position of the responsible genetic variant is located on the chromosome, their genetic linkage can be examined.”
“We chose barley for our study since it is one of the most important cereal crops that has been subjected to extensive genomic study and has abundant genomic and phenotypic data available.”
The study was published in Plant Phenomics.
Prof. Li and his team analyzed the phenotypic data for plant height and seed weight along with the high-density genome-wide SNP dataset for 12,828 barley samples, which included both domesticated and wild barley. Prior to employing GWAS analysis to identify SNPs linked to either of these variables, they first assessed the heritability and correlation of both traits.
Thereafter, they determined the pleiotropic effect (a mechanism for trait correlation where a genetic variant influences two or more phenotypic traits) of each SNP on plant height and seed weight using genomic structural equation modeling. Additionally, they evaluated the effect of genetic linkage on the two traits using linkage disequilibrium (LD) decay.
While plant height and seed weight were heritable and positively associated in domesticated barley, they were not noticeable in wild barley, the researchers discovered. Also, compared to wild barley, domesticated barley crops are shorter and have smaller seeds, which shows that artificial selection has had less of an impact on the scaling of plant height and seed weight.
Also, the team discovered 20 SNPs that were linked to at least three functional genes involved in plant growth and development and had pleiotropic effects on both plant height and seed weight.
Last but not least, the LD decay analysis revealed that the chromosome has a sizable number of genetic markers that affect one or both attributes. These findings unequivocally demonstrate the existence of two separate genetic pathways that underlie the scaling of plant height and seed weight in barley pleiotropy and genetic linkage.
“Our findings provide a better understanding of the genetic factors underlying size scaling, which, in turn, could expand the scope for exploring other mechanisms underlying the allometric scaling in plants. While our study examined size scaling in a single crop species, it points towards the possible existence of similar genomic mechanisms for allometric scaling across other plant species,” remarks Prof. Li.
The findings of this study may also have consequences for crop development where correlated qualities are wanted in the other direction because size scaling in barley is positively correlated, genetically limited, and minimally influenced by domestication.