Researchers have created a new neural network computational model that aids in the understanding of the brain’s complexity. To gain a better understanding of how a complex organ like the brain works, scientists strive to accurately understand both its detailed cellular architecture and the intercellular communications that occur within it.
Dr. Md. Abul Hassan Samee and his colleagues at Baylor College of Medicine have made significant strides in this direction. They developed advanced computational methods that led to new insights into the complexities of brain structure and function, potentially improving our understanding of this complex organ in both health and disease.
“At the moment, we have technologies that allow us to identify and locate individual cells within a tissue.” “We can also determine what products are produced by each individual cell in that tissue,” said corresponding author Samee, an assistant professor of molecular physiology and biophysics at Baylor and a member of the Center for Organ Repair and Renewal.
Mammalian brains are complex, with millions to hundreds of billions of cells, and when analyzed, they produce massive amounts of data. The challenge has been to find ways to integrate the information in those datasets in order to create a model that accurately reflects how an organ works.”
At the moment, we have technologies that allow us to identify and locate individual cells within a tissue. We can also determine what products are produced by each individual cell in that tissue.
Dr. Md. Abul Hassan Samee
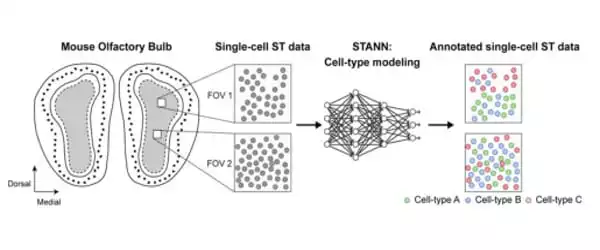
Samee worked with Francisco José Grisanti Canozo, the study’s first author, and Dr. James Martin, both of Baylor and the Texas Heart Institute, as well as Zhen Zuo, also of Baylor, to develop a neural network model to elucidate architectural and functional aspects of complex tissues. The model is known as Spatial Transcriptomics Cell-Type Assignment Using Neural Networks (STANN).
“We also used other advanced, sophisticated computational methodologies that strengthen the model,” Samee explained. “We applied STANN and other methods to existing mouse olfactory bulb brain datasets and began to see very interesting patterns in the brain’s cellular architecture and functionality.”
STANN enabled the researchers to predict a detailed picture of the brain’s cellular organization, as the brain has distinct morphological layers. “Layer by layer, our model provided the precise location of various cell types, whether they communicated with each other, and how,” Samee explained. “For us, this was a ‘eureka moment.'”
Samee and his colleagues discovered that the composition of cell types is quite consistent within a morphological layer. For example, a specific layer may have a constant percentage of astrocytes, neurons, and microglia throughout the layer. “These percentages look very similar if we take several small sections from different areas of the same morphological layer. However, the percentages may differ from one layer to the next “Samee elaborated.
When the researchers looked at cell colocalization, they noticed a different pattern emerge. “For example, astrocytes and olfactory neurons may colocalize in one area of a morphological layer. However, these cells can be completely separated in another area of the same morphological layer “Samee elaborated. “We also see that intercellular communication between two cell types varies in different areas of a morphological layer, indicating that gene regulatory networks vary with location.”
The researchers hypothesize that different spatially localized communities of cell types exist in the brain’s morphological layers. The cell types in the communities are similar, but there are significant differences in how cell types colocalize and communicate within a community. This suggests that brain cell types have spatially localized subtypes performing location-specific functions.
“This new detailed view of brain organization at the single-cell and functional levels had not previously been described,” Samee explained.
“The neural network model approach we developed in this work presents an ‘instruction manual’ for other researchers to use to study other areas of the brain or other organs, such as the heart, which is the main focus of my lab,” said co-author Martin, vice chairman and professor of molecular physiology and biophysics, Vivian L. Smith Chair in Regenerative Medicine, and director of the Center for Organ Repair and Renewal at Baylor College of Medicine. Martin is also the director of the Texas Heart Institute’s Cardiomyocyte Renewal Laboratory.