Nanopore technology has the potential to enable the development of small, portable, low-cost devices capable of real-time DNA sequencing. However, one of the challenges has been to improve the technology’s accuracy.
Any chemical, enzymatic, or technological procedure for determining the linear order of nucleotide bases in DNA is referred to as DNA sequencing. Sanger sequencing in the presence of dideoxynucleotide chain terminator monomers has now given way to ‘next-generation’ sequencing via short parallel read technologies.
Researchers at The University of Texas at Dallas have made strides toward this goal by developing a nanopore sequencing platform that can detect the presence of nucleobases, the building blocks of DNA and RNA, for the first time. The study was published and appears on the back cover of the print edition of Electrophoresis.
Researchers have moved closer toward this goal by developing a nanopore sequencing platform that, for the first time, can detect the presence of nucleobases, the building blocks of DNA and RNA.
“Our platform can help improve the sensitivity of nanopore sequencing by allowing us to detect the presence of nucleobases,” said Dr. Moon Kim, professor of materials science and engineering, and the Louis Beecherl Jr. Distinguished Professor in the Erik Jonsson School of Engineering and Computer Science.
Most DNA sequencing is currently done in the lab using fluorescent dye and lasers to determine the sequence of the four nucleobases, the fundamental units of the genetic code: adenine (A), cytosine (C), guanine (G), and thymine (T). When illuminated, each nucleobase emits a different wavelength, allowing scientists to determine the sequence.
The demand for sequencing has never been greater, with applications ranging from comparative genomics and evolution to forensics, epidemiology, and applied medicine for diagnostics and therapeutics. The pursuit of identifying and interpreting human sequence variation as it relates to health and disease is arguably the strongest rationale for ongoing sequencing.
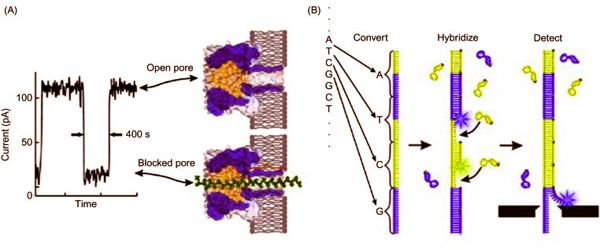
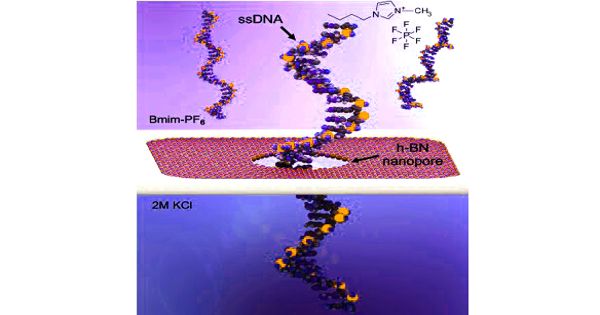
Sequence variation studies, comparative genomics and evolution, forensics, and diagnostic and applied therapeutics are just a few of the applications. Several emerging technologies show promise in terms of providing next-generation solutions for fast and affordable genome sequencing. A DNA sample is uncoiled and the hairlike strand is fed through a tiny hole, or nanopore, typically in a fabricated membrane, in nanopore sequencing. The DNA strand disrupts the electrical current flowing through the membrane as it moves through the nanopore. The current behaves differently depending on the properties of a DNA molecule, such as its size and shape.
“The electrical signal changes as the DNA moves through the nanopore,” Kim said. “We can read the characteristics of the DNA by monitoring the signal.”
In the field of Next Generation Sequencing, remarkable progress has been made (NGS). Parallelization of the sequencing reaction has greatly increased the total number of produced sequence reads per run due to advances in molecular biology and technical engineering. In molecular biology, sequencing is used to study genomes and the proteins they encode. Researchers can use sequencing data to identify changes in genes, associations with diseases and phenotypes, and potential drug targets.
The difficulty in controlling the speed of the DNA strand as it moves through the nanopore has been one of the challenges in advancing nanopore sequencing. The UT Dallas team addressed this by creating an atomically thin solid-state – or nonbiological – membrane coated with titanium dioxide, water, and an ionic liquid to slow the speed of molecules through the membrane. Water was added to the liquid solution to increase the amplitude of the electrical signals, making them easier to read.
“By enabling us to detect the presence of nucleobases, our platform can help improve the sensitivity of nanopore sequencing.”
The next step for researchers will be to improve the platform’s ability to quickly identify each nucleobase. According to Kim, the platform also allows for the sequencing of other biomolecules.
“The ultimate goal is to have a hand-held DNA sequencing device that is fast, accurate, and portable,” Kim explained. “This would lower the cost of DNA sequencing and make it more widely available.”