RNA-based antibiotics, like coronavirus vaccines, have the potential to significantly improve modern medicine. Teams of researchers have investigated the requirements that such antibiotics must meet in order for this strategy to work.
mRNA-based vaccines have shown impressive promise in the fight against the Covid-19 pandemic. Using this technology, scientists were able to rapidly develop and bring to market vaccines against SARS-CoV-2, which have proven to be extremely effective in protecting millions of people from severe COVID-19 disease progression or even death.
RNA-based medicine, on the other hand, can be used to combat more than just viruses. It can also be considered a candidate for a new type of antibiotic that can be used to treat bacterial infections in a tailored manner, among other things.
Researchers from the University of Würzburg investigated which prerequisites these active agents must meet and how they work in the bacterium (JMU). The Institute of Molecular Infection Biology (IMIB) and the Helmholtz Institute for RNA-based Infection Research (HIRI) collaborated on the study. They present their findings in the current issue of Nucleic Acids Research.
The number of antibiotic-resistant bacterial strains is increasing worldwide, and conventional active agents are increasingly failing to treat them. As a result, novel drugs to combat these pathogens in a targeted and effective manner are desperately needed.
Professor Jörg Vogel
Traditional antibiotics are increasingly failing
“The number of antibiotic-resistant bacterial strains is increasing worldwide, and conventional active agents are increasingly failing to treat them. As a result, novel drugs to combat these pathogens in a targeted and effective manner are desperately needed.” This is how Professor Jörg Vogel describes the history of the recently published work. Vogel is the corresponding author of this study and the chair of Molecular Infection Biology I at JMU.
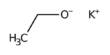
Antibiotics with programmable mRNA could be the answer to this problem. The strategy is straightforward: “We introduce short chains of bases into bacteria that are specifically designed to match specific genes,” Vogel explains. When these fragments bind to the corresponding mRNA of the gene of interest, they abrogate protein production, and, ideally, the bacterium dies as a result.
Switched off by the mirror image
In science, this approach is known as “antisense technology.” The structure of those active agents is a mirror image of a gene, enabling them to effectively block it. The first drugs that work via this principle are already on the market, such as those for treatment of the consequences of spinal muscular atrophy or hepatitis C infection. However, mRNA antibiotics have been confined to the laboratory so far.
Würzburg researchers focused their research on bacterial strains of the type “uropathogenic Escherichia coli (UPEC).” In the vast majority of cases, these bacteria cause a urinary tract infection in one out of every two women at some point in their lives. Antibiotic overuse in recent decades has resulted in the development of resistance to current therapeutics in many of these bacteria, complicating treatment of frequently occurring recurrent urinary tract infections.
Answers to three key questions
Three central questions were addressed by the research teams involved. First, are the active agents designed to be specific (particularly antisense peptide nucleic acids that target mRNAs of essential bacterial genes)? In other words, do they actually inhibit only one bacterial gene? Or could they be affecting other mRNAs as well? “Our results show that the applied base pairs only block the gene of interest,” Vogel says.
Second, how does the bacterium react to the RNA antibiotics being translocated into the cell? Bacteria exhibit a stress response, and thus, unfortunately, not as desired. This is primarily due to the relatively large size of antisense peptide nucleic acids. The stress therefore occurs primarily when these biomolecules cross the bacterial membrane.
However, there is good news with regard to the answer to question three: Is it possible to make these “base pair snippets” smaller? Yes, it is. “Until now, scientists have assumed that between nine and 14 base pairs are necessary to prevent any non-specific binding to other genes,” explains Vogel. The published results now show that nine base pairs are sufficient; thus, the snippets can be kept relatively small.
Overall, the study’s authors conclude that mRNA-based antibiotics are in principle suitable for combating uropathogenic Escherichia coli strains. However, several critical questions must be addressed before this approach can be used in clinics. There is still a pressing need: “If we don’t want antibiotic-resistant microbes to thwart modern medicine’s successes, we need novel tools that enable targeted treatment of pathogens,” says Jörg Vogel.