A crucial component in researching and developing economically significant crops is measuring plant phenotypes, a term used to describe the observable properties of an organism. Phenotypes important to breeding include characteristics like the number of kernels in corn, the size of the seeds in wheat, or the color of the grapes’ fruits.
Although these characteristics can be seen with the unaided eye, they are actually the result of microscopic molecular and cellular activities within the plant. A new development in the field of plant biology is the use of three-dimensional (3D) imaging to record phenotypes on a “whole-plant” scale, ranging from microscopic cells and organelles in the roots to leaves and flowers.
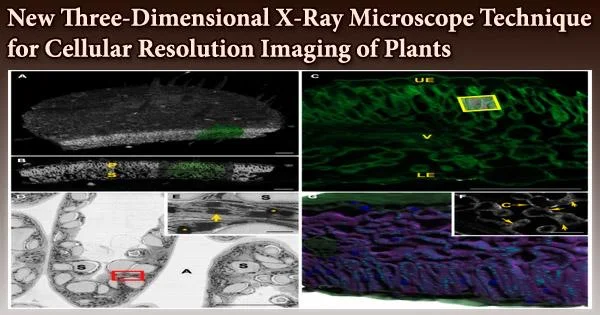
Current 3D imaging techniques can typically only image a few layers of cells within a plant tissue due to imaging depth limitations and laborious sample preparation. In recent studies, Keith Duncan, a research scientist in Christopher Topp’s lab, and others have developed X-ray microscope technology to image plant cells, whole tissues, and even organs at unprecedented depths with cellular resolution. Topp is an associate member at the Donald Danforth Plant Science Center.
The study, titled X-ray microscopy enables multiscale high-resolution 3D imaging of plant cells, tissues, and organs, was just published in the academic journal Plant Physiology with funding from Valent BioSciences LLC and Sumitomo Chemical Corporation. Plant scientists will be able to investigate above and below-ground features with unprecedented clarity thanks to this effort.
“This paper focuses on the multiscale,” says corresponding author Chris Topp, “because plants are multiscale. An ear of corn starts off as a microscopic group of cells called a meristem. Meristem cells will eventually form all the visible parts of the corn plant through division and growth.”
Plant roots drive a lot of important biological processes; they feed microbes in the soil, and in return the plants get phosphorus and nitrogen. We know the interaction between roots and microbes is important because it was a primary source of phosphorus and nitrogen before we invented chemical fertilizers.
Christopher Topp
The researchers’ enhanced 3D X-ray microscopy (XRM) equipment enables them to link the plant’s developing microstructure, like its meristem cells, to its outward characteristics as it matures, including leaves and flowers.
To put it another way, 3D XRM offers cellular-level resolution of all the organs and tissues of the plant. Additionally, their XRM technique can photograph subterranean structures with outstanding resolution, such as roots, fungi, and other microorganisms.
“Plant roots drive a lot of important biological processes; they feed microbes in the soil, and in return the plants get phosphorus and nitrogen,” explains Topp. “We know the interaction between roots and microbes is important because it was a primary source of phosphorus and nitrogen before we invented chemical fertilizers.”
Our reliance on chemical fertilizers in common farming techniques has also significantly impacted the global environment.
“Half of all the biologically-available nitrogen was made in a factory in the last 100 years,” Topp continues. “This process has been estimated to use 3% of all available energy and generate 3% of greenhouse gas emissions on planet Earth every single year.”
Reducing chemical inputs and promoting natural interactions between roots and underground bacteria is therefore essential to the sustainable agriculture movement.
“We haven’t had the tools to understand these interactions until recently,” says Topp. “3D XRM can help unlock the potential of re-establishing these natural alliances in our agriculture systems.”
Due to its capacity to produce virtually flawless 3D clarity of plant structure, 3D XRM technology stands apart from other imaging methodologies in plant biology. Other popular techniques, including photon-based tomography, are constrained by modest imaging depths and are best used with a small number of plant species.
In contrast, by using 3D XRM, the team led by Topp and Duncan are able to image “thick tissues that are recalcitrant to typical, optical methods,” in a whole host of economically important crops, including corn, foxtail millet, soybean, teff, and grape. “This paper is the first of its kind to show the breadth of what 3D XRM can do,” Topp notes.
The establishment of a repeatable technique for other plant scientists interested in 3D XRM imaging is one of the main objectives of the paper. Lead author Keith Duncan worked hard and iteratively to prepare samples in order to maximize the contrast between the plant and its surroundings.
Due to differential absorption, which is how X-ray imaging operates, dense material (such as soil minerals) absorbs more X-rays and appears darker on an image. The researchers was in danger of totally wiping out the material they were interested in imaging since living substance, such as plant tissue, has low X-ray absorption.
“Solving that problem for one kind of sample like a root tip is one thing,” explains Topp, “but the idea of the paper was to give plant scientists working on a variety of relevant plant tissues and species the access to these methods. We want to broadly apply 3D XRM to plant systems above and below ground.”
As a result, the number of plant species and the kinds of plant tissues that can be photographed at almost perfect resolution have substantially increased thanks to their published methodology.
Keith Duncan is still in charge of the Topp Lab’s collaboration with Valent Biosciences and Sumitomo Chemical, which is aimed at enhancing 3D XRM capabilities. He frequently works with Kirk Czymmek, PhD, head of the Advanced Bioimaging Laboratory at the Danforth Center, who was also a co-author on the study.
Imaging 3D architecture of microbial networks in the soil is the next task on the horizon. A portion of the work involves enhancing machine learning techniques so that a computer may be trained to identify what is a root, soil, or spore (the reproductive cells of a fungus) inside a picture.
From the microscopic to the visible, their effort will continue to develop new technology methods to enhance our understanding of the “whole plant” on various scales.