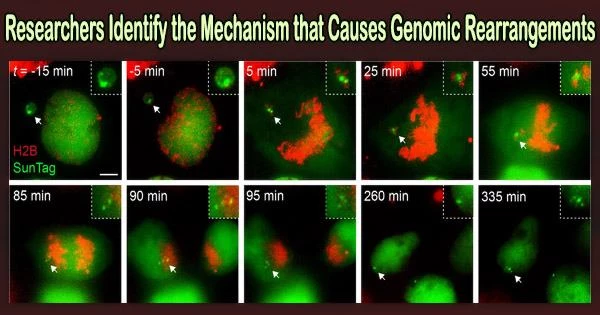
Every cell that divides aims to precisely divide its genome into two daughter cells that are genetically identical to it. Scientists from the UT Southwestern Medical Center say in a recent study that this process frequently goes wrong and may be the cause of a new class of chromosomal defects identified in malignancies and congenital diseases.
The discovery, published in Nature, sheds light on how cancer cells rapidly evolve genomic changes that fuel their proliferation.
“Cancer genomes are remarkably complex. Our findings provide a fundamental understanding of how diverse patterns of chromosomal alterations form and drive cancer development,” said Peter Ly, Ph.D., Assistant Professor of Pathology and Cell Biology at UT Southwestern, who co-led the study with Yu-Fen Lin, Ph.D., Senior Research Scientist.
The term “chromothripsis,” which is borrowed from Greek and refers to the catastrophic breaking of chromosomes into tiny fragments, is the main topic of the essay.
In the study, Dr. Ly and associates in the Ly lab looked into the movement of broken chromosomes from aberrant micronuclei during cell division. The scientists discovered that during cell division, the chromosomal pieces did not disperse throughout the cell but instead stayed bound together.
These findings suggest that chromosomally unstable and/or DNA repair-deficient tumors with micronuclei may be vulnerable to CIP2A inhibition.
Dr. Peter Ly
This made it possible for the fragmented chromosome to enter one of the two daughter cells as a whole. There, the cell’s DNA repair mechanism randomly put the fragments back together in the wrong order to create a rearranged chromosome.
The researchers identified a protein complex consisting of CIP2A and TOPBP1 that tethers the DNA fragments together during cell division. The loss of important tumor suppressor genes is caused by the genetic signatures of this process, which are seen in 25 different cancer types.
These findings build upon previous work by Dr. Ly, who has engineered unique experimental systems to re-create and study chromothripsis in the laboratory.
“Since the discovery of chromothripsis in cancer genomes, it was puzzling to understand how fragments of a shattered chromosome could reassemble to generate scrambled chromosomes. We have now solved a key piece of that puzzle,” said Dr. Ly, a member of the Harold C. Simmons Comprehensive Cancer Center and a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar in Cancer Research.
The chromosome fragments scattered during cell division when the researchers prevented tethering by deleting the gene encoding for CIP2A or TOPBP1. This caused them to amass throughout the nucleus and cytoplasm of both daughter cells, ultimately killing both.
“These findings suggest that chromosomally unstable and/or DNA repair-deficient tumors with micronuclei may be vulnerable to CIP2A inhibition,” said Dr. Ly. His lab plans to continue studying the role of CIP2A-TOPBP1 in maintaining genome stability and whether this protein complex could be a relevant target for cancer treatment.
Other UT Southwestern researchers who contributed to this study are Qing Hu, Ph.D., Alice Mazzagatti, Ph.D., and Rashmi Dahiya, Ph.D., Postdoctoral Fellows; and Elizabeth Maurais, Justin Engel, and Giaochau Nguyen, students in the Graduate School of Biomedical Sciences.