Molecular monitoring of RNA regulation involves techniques that allow researchers to study how RNA (ribonucleic acid) is regulated within cells. This can include techniques such as RNA sequencing, which allows researchers to determine the sequence of RNA molecules and how they are transcribed from DNA, as well as techniques such as RT-qPCR (reverse transcriptase-quantitative polymerase chain reaction) which can be used to quantify the expression levels of specific RNA molecules in a sample.
Other techniques that can be used for molecular monitoring of RNA regulation include Northern blotting, in situ hybridization, and microarrays. These techniques can provide insight into the role that RNA plays in gene expression and regulation, and can be useful in a variety of research and clinical applications.
Molecular biology methods have enormous value not only in the investigation of basic scientific questions, but also in the application to a wide range of problems affecting the overall human condition. Disease prevention and treatment, the creation of new protein products, and the manipulation of plants and animals for desired phenotypic traits are all applications that are routinely addressed by the use of molecular biology methods.
The better we understand cellular processes like RNA regulation, the better molecular therapies we can develop. Until now, it has been especially difficult to track the regulation of non-coding RNA, which is RNA that is not further converted into proteins. A research team from Helmholtz Munich and the Technical University of Munich (TUM) has now developed a minimally invasive reporter system that enables highly sensitive monitoring of RNA production of both coding and non-coding RNA.
For cellular processes, our genetic DNA information is transcribed into RNA, which is then processed further before serving as a blueprint for proteins or performing a cellular function on its own. The types of RNA produced and the quantities produced reveal a lot about the health of our cells. Cells, for example, produce more RNA molecules that code for proteins involved in the immune response during an infection.
Researchers can use existing reporter systems to track the process of DNA molecules being translated into proteins via RNA. Not all human genes, however, encode proteins. The vast majority of human genes are non-coding, including those encoding long non-coding RNAs (lncRNA). These are RNA molecules with more than 200 building blocks that do not act as blueprints for proteins. Instead, they control important processes in cells. Initial research shows that lncRNA is involved in such processes as regulating RNA production, the organization of structures in the cell nucleus or in switching certain enzymes on and off.
Despite their importance for cellular processes, it has been difficult to investigate lncRNAs with existing methods. So far, this was only partially possible, for example in fixed cells at specific time points, because classical reporter systems based on the translation into proteins cannot be used.
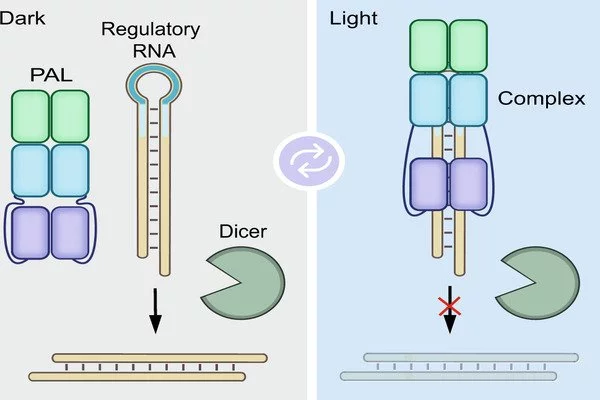
INSPECT, unlike previous methods, encodes sequences for reporter proteins in modified introns. These are sequences in the pre-mature RNA molecule that are naturally removed and eliminated by the cell during processing.
Dong-Jiunn Jeffery Truong
INSPECT permits the monitoring of non-coding RNA
INSPECT, a new reporter system, has been developed as a solution. The newly developed reporter system was published in Nature Cell Biology by a team led by Gil Westmeyer, Professor of Neurobiological Engineering at TUM and Director of the Institute for Synthetic Biomedicine at Helmholtz Munich.
“INSPECT, unlike previous methods, encodes sequences for reporter proteins in modified introns. These are sequences in the pre-mature RNA molecule that are naturally removed and eliminated by the cell during processing. INSPECT stabilizes introns so that, rather than being degraded after removal, they are transported to the cellular cytoplasm and translated into reporter proteins” Dong-Jiunn Jeffery Truong, the first author, elaborates. The researchers can then use conventional methods to detect reporter protein signals such as fluorescence.
INSPECT modifies neither the completed RNA nor the proteins
Thus, the new molecular biology tool not only solves the problem of tracking the generation of non-coding RNA, but also provides advantages for studying coding RNA. Current reporter systems frequently risk damaging the RNA or proteins under investigation, for example, because they must be fused directly to the RNA being studied in order to be co-translated into proteins. INSPECT modifies introns rather than the completed RNA or proteins.
The team demonstrated the function of INSPECT using various examples of coding and non-coding RNA. They monitored the production of RNA for interleukin 2, a protein that is produced in greater quantities in response to infections. They have also achieved highly sensitive monitoring of the production of two lncRNAs and tracked changes in regulation during the investigation period.
“INSPECT adds an important molecular biology tool to the biomedical toolbox. It makes it easier to study the role of certain non-coding RNA molecules in cell development and to investigate how their regulation can be modulated, for example, to prevent cancer cells from forming “Prof. Westmeyer explains. “With the minimally invasive reporter system EXSISERS, which we previously developed to study protein isoforms, it may be possible in the future to study an entire genetic regulation process in living cells, from RNA processing to the production of specific protein variants.”