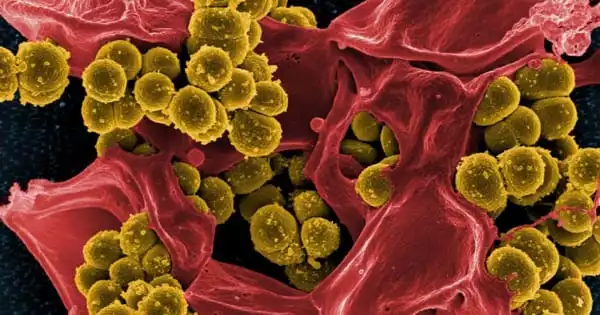
Superbugs are microorganisms that have developed resistance to the treatments that should kill them. These drug-resistant bacteria and fungi are difficult to control and treat. Superbugs are germs that have developed resistance to antibiotics. They could possibly be fungus. Antibiotics are a critical class of medications that help save many lives. They treat a wide range of infections, from minor urinary tract infections to life-threatening sepsis. However, the recent rise in superbugs is partly due to antibiotic abuse, which adds to antibiotic resistance.
Methicillin-resistant Staphylococcus aureus (MRSA) is a bacterial infection that has developed resistance to most antibiotics used to treat common staph infections. Bruce Donald, a computer scientist at Duke, and colleagues at the University of Connecticut are seeking to create novel enzyme inhibitors to combat MRSA. The researchers revealed how a single tiny mutation affects therapeutic efficacy in a study published in PLOS Computational Biology.
They looked at dihydrofolate reductase (DHFR), an enzyme that drugs use to combat MRSA. Drugs that inhibit DHFR function similarly to locks and keys in that they bind to MRSA enzymes, which have a particular three-dimensional structure that only permits molecules that fit precisely to attach to them.
We looked at the electron density data from the crystallographer and discovered something odd. Crystallographers utilized a computer program that, unbeknownst to them, flipped the chirality, or made a mirror image, of the NADPH cofactor to gain a better fit when attempting to determine the structure of the F98Y mutant.
Pablo Gainza
A mutation can alter the structure of a bacterial enzyme, causing medications to lose their potency. The F98Y mutation is a well-known resistance mutation. A little alteration in the 98th amino acid in the DHFR enzyme converts phenylalanine to tyrosine. “Those two amino acids are structurally similar,” said Graham Holt, a grad student in the Donald lab, “yet the mutation has a big effect on the efficacy of the inhibitors.” In essence, it modifies the lock.
Pablo Gainza, PhD, a former graduate student in the Donald lab, believed he could forecast this mutation using OSPREY, a suite of algorithms developed in the Donald lab for computational structure-based protein design. But he couldn’t do it. He returned to the original structure after rejecting hypothesis after hypothesis to figure out why he couldn’t predict this mutation.
“We looked at the electron density data from the crystallographer and discovered something odd,” Donald explained. Crystallographers utilized a computer program that, unbeknownst to them, flipped the chirality, or made a mirror image, of the NADPH cofactor to gain a better fit when attempting to determine the structure of the F98Y mutant. The “flipped” chemical species they uncovered in their analysis exists in laboratory circumstances and maybe in vivo.
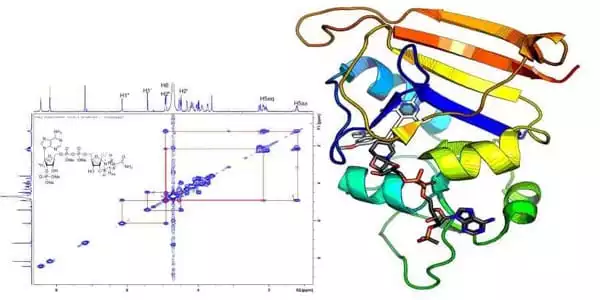
“We detected this inverted chirality using OSPREY,” Donald explained, “which we assume occurred as a result of the F98Y mutation.” The single enzyme mutation and the flipped cofactor appear to work together to circumvent the inhibitor, as in 2-factor authentication.
The fundamental basis for resistance is altered by this “chiral evasion.” However, Donald and colleagues now understand not only how a single little mutation changed the lock, but also the structure required to create a better key – a better pharmacological inhibitor.
“This is the first example of an enzyme that uses the chirality of its cofactor to avoid inhibition,” Holt added. “Now that we’re seeing this, it’ll help inform computational tactics for developing better inhibitors.”
The Donald lab demonstrated that by accounting for flipped chirality, OSPREY’s predictions closely match experimental measurements of inhibitor potency. They collaborated with researchers at the University of Connecticut on biochemical studies to test the notion and give structural support.
“This is just the start of the story,” Donald explained. “Our discovery of chiral evasion should lead to more resilient inhibitors, resulting in better therapeutic designs.” Most drug design is currently reactive, waiting for resistance to emerge, which it invariably does. “By employing our algorithms to anticipate resistance, we hope to make drug design proactive,” Donald said.
According to the Centers for Disease Control and Prevention, antibiotic-resistant illnesses cause more than 35,000 fatalities in the United States each year (CDC). Antibiotic-resistant bacteria may be more prevalent in areas that must be sterilized on a regular basis, such as hospitals and other healthcare institutions. Regular sterilization is critical for preventing infections in these environments, but it may also be strengthening some microorganisms. Furthermore, these harmful microorganisms may be more prevalent in specific meals, such as animal products treated with antibiotics by farmers.