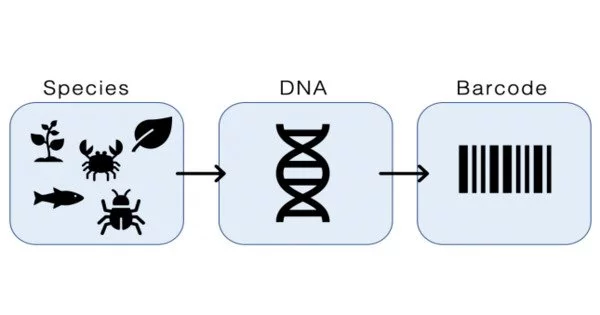
Microbial DNA barcoding is a scientific approach that uses unique DNA sequences to detect and classify bacteria. It is the use of DNA metabarcoding to characterize a microbial combination. It is a useful tool for taxonomic and biodiversity research, as well as for environmental microbiology, microbial ecology, and clinical microbiology.
DNA metabarcoding is a DNA barcoding technology that uses universal genetic markers to identify DNA from a variety of organisms. The word “barcoding” was coined in the field of genetics to refer to a short, standardized DNA sequence used to identify species.
Here’s how microbial DNA barcoding works:
- Target Gene Selection: A specific gene or genomic area is chosen as a barcode in microbial DNA barcoding. This gene is often preserved within a certain group of microorganisms but differs between taxa, making it useful for identification. The barcode gene is chosen based on the group of microorganisms being researched.
- DNA Extraction: DNA is extracted from the microbiological sample of interest, which could be soil, water, or air, clinical specimens (e.g., swabs, sputum), or cultured isolates.
- PCR Amplification: Polymerase Chain Reaction (PCR) is used to amplify the target gene from extracted DNA. In this phase, primers designed to precisely target the specified barcode gene are utilized.
- Sequencing: The amplified DNA is then sequenced using modern DNA sequencing techniques, such as Sanger sequencing or next-generation sequencing (NGS). The resulting sequence data provides the genetic information required for identification.
- Data Analysis: After obtaining the sequences, bioinformatics tools and databases are used to compare the obtained DNA sequence with reference sequences in databases (e.g., GenBank, SILVA) or with a reference dataset developed particularly for the barcode gene of interest. This comparison allows the microorganism to be assigned to a certain taxonomic level, such as species or genus.
Applications
Many studies have followed Woese et al.’s first application and are now covering a wide range of applications. Metabarcoding is applied in fields other than biology and ecology. Bacterial barcodes are also employed in medicine and human biology, for example, to explore the microbiome and bacterial colonization of the human gut in normal and obese twins, or in comparison studies of newborn, child, and adult gut bacteria composition.
The 16S rRNA gene for bacteria and archaea, the 18S rRNA gene for eukaryotic microorganisms like fungus and protists, and the internal transcribed spacer (ITS) sections for fungi are all commonly used barcode genes for microbial DNA barcoding. These genes have changeable sections that can be used to discriminate between various species or taxa.
Microbial DNA barcoding is a potent approach for describing and studying microbial populations with applications in environmental research, agriculture, biotechnology, and medicine.