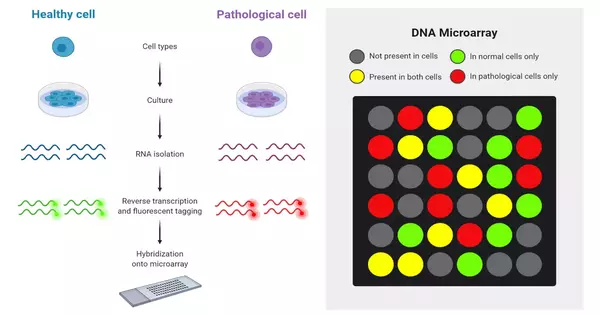
Microarray analysis is a powerful technique in molecular biology and genomics that allows researchers to study the expression of thousands of genes at the same time. Microarray analysis techniques are used to interpret data from DNA (Gene chip analysis), RNA, and protein microarray experiments, which allow researchers to investigate the expression state of a large number of genes – in many cases, an organism’s entire genome – in a single experiment.
These experiments can produce massive amounts of data, allowing researchers to assess the overall state of a cell or organism. It enables researchers to examine gene expression levels under various conditions, assisting in the understanding of gene regulation and the identification of potential biomarkers for various diseases. Without the assistance of computer programs, analyzing data in such large quantities is difficult, if not impossible. Here is an overview of the basic steps involved in microarray analysis:
Experimental Design:
- Define the research question and objectives.
- Choose appropriate biological samples and experimental conditions.
- Consider the type of microarray platform (e.g., DNA microarray or gene expression microarray) based on the study goals.
RNA Extraction and Labeling:
- Isolate RNA from the biological samples (usually total RNA).
- Purify and assess the quality of RNA.
- Reverse transcribe RNA into complementary DNA (cDNA).
- Label cDNA with fluorescent dyes (e.g., Cy3 and Cy5) for dual-color microarrays.
Hybridization:
- Mix labeled cDNA from experimental and control samples.
- Apply the mixture to the microarray slide or chip containing immobilized probes (usually DNA sequences representing genes).
- Incubate the slide to allow hybridization between labeled cDNA and immobilized probes.
Scanning and Data Preprocessing:
- Use a microarray scanner to detect fluorescence signals from labeled cDNA on the microarray.
- Obtain raw intensity data for each spot on the microarray.
- Normalize the raw data to correct for systematic variations (e.g., background noise, dye bias) between samples.
- Filter out low-quality spots and perform background correction.
Data Analysis:
- Perform statistical analysis to identify differentially expressed genes between experimental and control groups.
- Apply fold-change and p-value thresholds to prioritize significant genes.
- Use bioinformatics tools for functional annotation, pathway analysis, and clustering to interpret the biological significance of the results.
Microarray analysis has progressed, and next-generation sequencing (NGS) technologies such as RNA-Seq are increasingly being used to study gene expression. Microarrays, on the other hand, remain useful for certain applications and can provide cost-effective solutions for large-scale gene expression profiling.