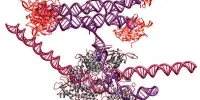
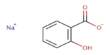
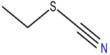
A thorough investigation of macroalgae genetics has shown the genetic foundations that allowed macroalgae, or “seaweed,” to evolve multicellularity. Researchers in the journal Molecular Plant describe that three lineages of macroalgae achieved multicellularity independently and during very diverse time periods by gaining genes that allow for cell adhesion, extracellular matrix creation, and cell differentiation. Surprisingly, many of these multicellular-enabling genes originated in viruses. The study, which increased the total number of sequenced macroalgal genomes from 14 to 124, is the first to look at macroalgal evolution from a genomic perspective.
“This is a big genomic resource that will open the door for many more studies,” says co-first author and algal biologist Alexandra Mystikou of New York University Abu Dhabi and the Technology Innovation Institute, United Arab Emirates. “Macroalgae play an important role in global climate regulation and ecosystems, and they have numerous commercial and ecoengineering applications, but until now, there wasn’t a lot of information about their genomes.”
Macroalgae play an important role in global climate regulation and ecosystems, and they have numerous commercial and ecoengineering applications, but until now, there hasn’t a lot of information about their genomes.
Alexandra Mystikou
Macroalgae thrive in both fresh and salt water and are complex multicellular organisms with separate organs and tissues, as opposed to microalgae, which are minuscule and unicellular. Macroalgae are classified into three groups: red (Rhodophyta), green (Chlorophyta), and brown (Ochrophyta), each of which evolved multicellularity at a different epoch and under different environmental conditions. Rhodophytes and Chlorophytes gained multicellularity more than a billion years ago, whereas Ochrophytes only became multicellular in the last 200,000 years.
To explore the evolution of macroalgal multicellularity, the researchers sequenced 110 additional macroalgal genomes from 105 different species found in fresh and saltwater habitats across various geographies and temperature zones.
The researchers discovered multiple metabolic pathways that distinguish macroalgae from microalgae, some of which may explain the success of invading macroalgal species. Many of these metabolic genes appear to have been supplied by algal-infecting viruses, and virally derived genes were particularly common in more recently evolved brown algae.
They discovered that macroalgae gained numerous novel genes that microalgae did not have on their path to multicellularity. For all three lineages, major acquisitions included genes involved in cell adhesion (which allows cells to stick together), cell differentiation (which allows different cells to develop specialized tasks), cell communication, and inter-cellular transport.
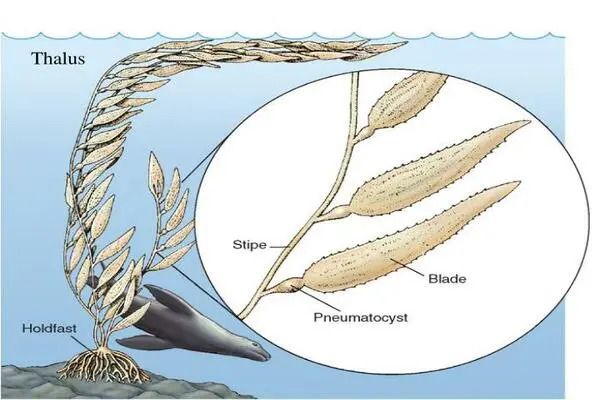
“Many brown algal genes associated with multicellular functions had signature motifs that were only otherwise present in the viruses that infect them,” says co-first author and bioinformatician David Nelson of New York University Abu Dhabi. “It’s kind of a wild theory that’s only been hinted at in the past, but from our data it looks like these horizontally transferred genes were critical factors for evolving multicellularity in the brown algae.”
The team also identified other features that were distinct between the macroalgal lineages. They observed much more diversity between different species of Rhodophyte, which evolved multicellularity first and have thus had longer to diverge. They also found that Chlorophytes share many genomic features with land plants, suggesting that these genes may have already been present in the last common ancestor of Chlorophytes and plants.
“By no means have we exhaustively explored all that there is in these genomes,” says senior author and systems biologist Kourosh Salehi-Ashtiani of New York University Abu Dhabi. “There is a ton of information that we have not touched in the present paper that can be mined by whoever who is interested.”
The researchers have already begun digging into the dataset to explore macroalgae’s environmental and habitat adaptations. In the future, they plan to sequence and analyze more macroalgal genomes.
“We want to explore some of these features in more detail, meaning more genomes if we can get our hands on them,” said Salehi-Ashtiani.