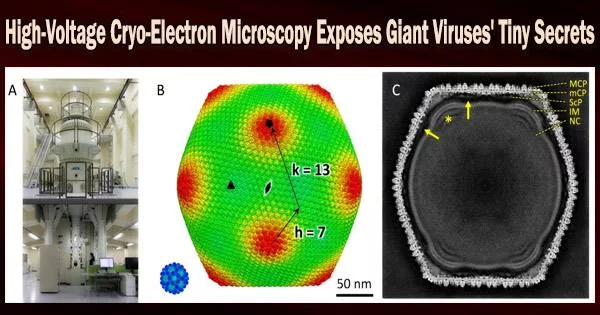
Giant viruses, despite their moniker, are challenging to visualize in detail. They are both too large and too little to be studied with optical microscopy, which is often employed to study larger specimens.
With the use of cryo-high-voltage electron microscopy, an international cooperation has now for the first time revealed the structure of tokyovirus, a large virus named for the city in which it was found in 2016.
They published their results on Dec. 12 in Scientific Reports.
“Giant viruses are exceptionally large physical size viruses, larger than small bacteria, with a much larger genome than other viruses,” said co-corresponding author Kazuyoshi Murata, project professor, Exploratory Research Center on Life and Living Systems (ExCELLS) and National Institute for Physiological Sciences, the National Institutes of Natural Sciences in Japan.
“Few studies have revealed the capsid the protein shell encapsulating the double-stranded viral DNA structure of large icosahedral, or 20-sided, viruses in detail. They present special challenges for high-resolution cryo-electron microscopy from their size, which imposes hard limits on data acquisition.”
The researchers employed one of the few high-voltage electron microscopy (HVEM) facilities in the world that is capable of imaging biological material to overcome the difficulty. This kind of electron microscope accelerates voltage to, in theory, boost the power of the microscope and enable higher-resolution imaging of thicker samples.
At the Research Center for Ultra-High Voltage Electron Microscopy at Osaka University, the team imaged flash-frozen tokyovirus particles, with the goal of reconstructing a single particle in full detail for the first time.
Giant viruses are exceptionally large physical size viruses, larger than small bacteria, with a much larger genome than other viruses. Few studies have revealed the capsid the protein shell encapsulating the double-stranded viral DNA structure of large icosahedral, or 20-sided, viruses in detail. They present special challenges for high-resolution cryo-electron microscopy from their size, which imposes hard limits on data acquisition.
Professor Kazuyoshi Murata
“Cryo-HVEM on biological samples has not been previously reported for single particle analysis,” Murata said. “For thick samples, such as tokyovirus with a maximum diameter for 250 nanometers, the influence of the depth of field causes an internal focus shift, imposing a hard limit on attainable resolution. Accelerating the voltage, or shortening the wavelength of the emitted electrons, can increase the depth of field and improve the optical conditions in thick samples.”
The researchers scanned the tokyovirus in great detail to elucidate the structure of the entire virus particle after making these changes. They were able to recreate a 3D image at a resolution of 7.7 angstroms, which is only marginally less than what the technology is capable of. According to Murata, the resolution’s outcome was constrained by the amount of data they could gather.
“Cryo-HVEM currently requires the manual collection of micrographs taken with the microscope,” Murata said. Micrographs are photographs taken with the microscope. “We identified 1,182 particles from 160 micrographs, which is an extremely small number compared to reports of other giant viruses imaged with less powerful microscopes.”
Murata asserts that while a lower magnification expands the number of particles seen in each micrograph, a high enough magnification is required to picture the particles clearly.
The number of images taken at high magnification has increased significantly thanks to the automated acquisition of micrographs used in standard cryo-electron microscopy, but the manual mode allowed researchers to maintain a better particle count per micrograph while also maintaining a higher sampling frequency.
“Even with limited samples and slightly lower resolution,” Murata said, “the researchers gathered enough information to better understand the giant virus particles structure with more clarity than ever before.”
“The cryo-HVEM map revealed a novel capsid protein network, which included a scaffold protein component network,” Murata said, noting that this scaffolding network’s connection between vertices in the icosahedral particle may determine the particle size.
“Icosahedral giant viruses, including tokyovirus, have large, uniform sized functional cages created with limited components to protect the viral genome and infect the host cell. We are beginning to learn how this works, including the advanced functions of the structures and how we might be able to apply this understanding.”
“The researchers plan to implement automated acquisition software capable of maintaining their desired parameters to image more giant virus structures and discover common architecture to better understand how the limited structures can be used for multifunctional organisms,” Murata said.
Murata is also affiliated with the Department of Physiological Sciences in the School of Life Science at The Graduate University for Advanced Studies (SOKENDAI). Other contributors include co-corresponding author Chihong Song, co-first author Akane Chihara and co-first author Raymond N. Burton-Smith, who are affiliated with ExCELLS and the National Institute for Physiological Sciences; Naoka Kajimura and Kaoru Mitsuoka, with the Research Center for Ultra-High Voltage Electron Microscopy, Osaka University; and Kenta Okamoto, Program in Molecular Biophysics, Department of Cell and Molecular Biology, Uppsala University in Sweden. Song and Chihara are also affiliated with SOKENDAI.
Japan’s Ministry of Education, Culture, Sports, Science and Technology; Joint Research of ExCells; Cooperative Study Program of National Institute for Physiological Sciences; SOKENDAI; The Swedish Research Council; Swedish Foundation for International Cooperation in Research and Higher Education; and Royal Swedish Academy of Sciences supported this research.