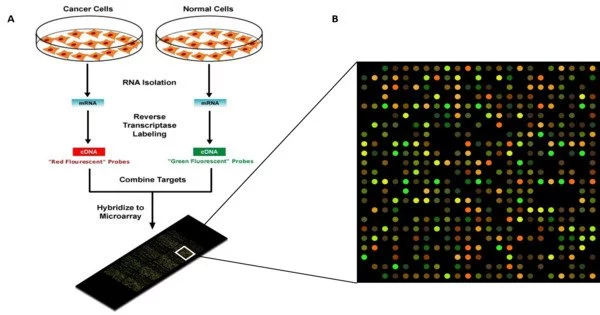
A DNA microarray is a grouping of microscopic DNA spots that are attached to a solid surface. A gene chip, also known as a DNA chip, is a technology that measures the activity of genes in a genome. DNA microarrays are used by scientists to simultaneously measure the expression levels of a large number of genes or to genotype multiple regions of a genome. It enables scientists to assess the expression levels of thousands of genes at the same time in a single experiment.
This technology has been especially useful in genomics and functional genomics, revealing gene expression patterns, gene regulation, and the molecular mechanisms underlying various biological processes.
Each DNA spot contains probes (or reporters or oligos), which are picomoles (10-12 moles) of a specific DNA sequence. These can be a short section of a gene or other DNA element used to hybridize a cDNA or cRNA (also known as anti-sense RNA) sample (called target) under stringent conditions.
Here’s a brief overview of how DNA microarrays work:
- Probe Design: Probes are short DNA sequences that are designed to be complementary to specific genes or gene variants. Typically, these probes are synthesized and immobilized on a solid surface, such as a glass slide or silicon wafer, to form an array.
- Sample Preparation: The fluorescent dye is used to label the target DNA or RNA after it has been extracted from cells or tissues. The labeled DNA or RNA is then hybridized to the microarray’s immobilized probes, forming complementary base pairs.
- Hybridization: The sample’s labeled DNA or RNA binds to its complementary probe on the microarray via base pairing. The amount of target DNA or RNA present in the sample is proportional to the degree of fluorescence at each spot on the array.
- Scanning and Analysis: The microarray is scanned to measure the fluorescence at each spot. The resulting data provide information about the expression levels of thousands of genes simultaneously. High fluorescence indicates high gene expression, while low fluorescence indicates low expression.
It was invented by Patrick O. Brown. An example of its application is in SNP arrays for polymorphisms in cardiovascular diseases, cancer, pathogens, and GWAS analysis. It is also used for the identification of structural variations and the measurement of gene expression.
Applications of DNA Microarrays:
- Gene Expression Profiling: Microarrays are widely used to study gene expression patterns in different tissues, developmental stages, or under various experimental conditions. This helps researchers identify genes involved in specific biological processes.
- Disease Diagnosis and Classification: DNA microarrays have been used to classify diseases based on gene expression profiles. This can aid in the diagnosis, prognosis, and treatment of various diseases, including cancer.
- Drug Discovery: Microarrays are important in drug discovery because they identify genes or pathways that are affected by potential drugs. This data assists researchers in understanding drug mechanisms of action and assessing potential side effects.
- Functional Genomics: Microarrays aid in the study of gene function by revealing how genes respond to environmental or experimental changes.
While DNA microarrays have been widely used, newer technologies such as RNA sequencing (RNA-Seq) have gained popularity for gene expression analysis due to their increased sensitivity and ability to detect novel transcripts. Despite this, DNA microarrays remain useful tools in genomics research.