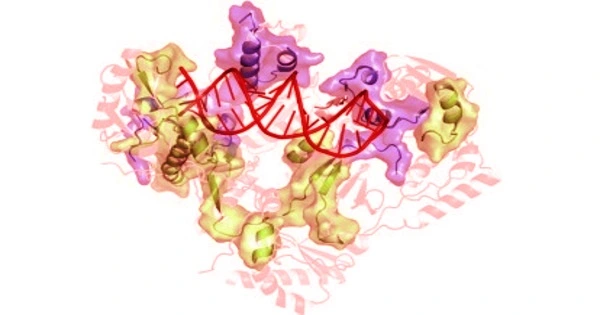
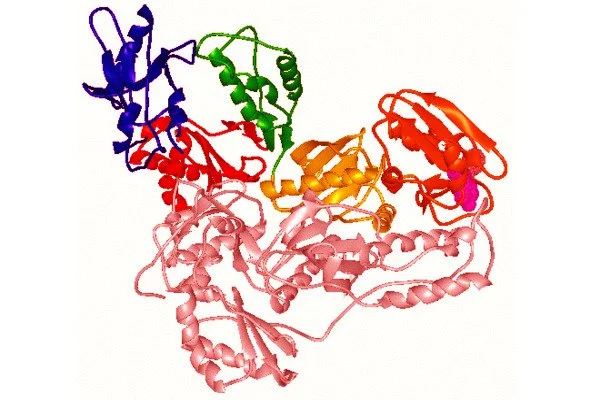
According to a study co-authored by a Rutgers researcher, the crystal structure of a human endogenous reverse transcriptase has similarities to HIV reverse transcriptase, a well-known tractable drug target, which will aid in the design of drugs to treat cancer and other diseases.
The study, which was published in the Proceedings of the National Academy of Sciences (PNAS), describes the first high-resolution three-dimensional structure of an endogenous reverse transcriptase, specifically human endogenous retrovirus-K (HERV-K) reverse transcriptase (RT). Previous research has discovered that a significant portion of the human genome is made up of repetitive elements that are relics of previous viral infections and are linked to a variety of serious diseases, including cancer.
Human endogenous retroviruses (HERVs) account for nearly 8% of the human genome and are derived from ancient retrovirus integrations into the germline. HERV-encoded genes produce functional proteins, including reverse transcriptases (RTs), which may contribute to the pathology caused by aberrant HERV-K expression.
Characterizing the structure of HIV RT was a critical turning point in designing novel medicines to combat that deadly virus. This study marks a significant step forward in our understanding of endogenous retroviruses and how they could be targeted to treat disease.
Eddy Arnold
The structure, according to the study, provides therapeutic opportunities for RT inhibitors-antiretroviral drugs used to treat HIV infection or AIDS, as well as hepatitis B-in cancer, autoimmune, and neurodegenerative diseases.
“This study marks a significant step forward in our understanding of endogenous retroviruses and how they could be targeted to treat disease,” said Eddy Arnold, resident faculty member at the Rutgers Center for Advanced Biotechnology and Medicine (CABM) and scientific advisory board member of biotechnology company ROME Therapeutics.
“Characterizing the structure of HIV RT was a critical turning point in designing novel medicines to combat that deadly virus,” said Arnold, a Distinguished Professor and Board of Governors Professor of chemistry and chemical biology at Rutgers. “Similarly, deeper insights into human endogenous RT could pave the way toward a new class of therapies for cancer and other serious diseases.”
Repetitive elements in the genome such as HERV-K are frequently overexpressed in cancer and elicit biological viral mimicry responses that can alter the tumor microenvironment, according to past research.
The abundant HERV-K family is distinct among the numerous endogenous retroviral elements found in the human genome because several members are transcriptionally active and code for biologically active proteins. Based on the partial sequence of the reverse transcriptase (RT) gene, a detailed phylogeny of the HERV-K family revealed a high incidence of an intact RT open reading frame within the HML-2 subgroup of HERV-K elements.
The study was co-authored by researchers from ROME Therapeutics, a biotechnology company that aims to develop novel therapies for cancer and autoimmune diseases by researching the Dark Genome for drug development (vast stretches of uncharted genetic material that account for more than 60% of the human genome).
“In this paper, we describe for the first time the crystal structure of an endogenous reverse transcriptase, known as HERV-K RT, and show that it has striking similarities to HIV reverse transcriptase, a well-known tractable drug target,” said Dennis Zaller, ROME’s chief scientific officer.
“This accomplishment is a watershed moment in the Dark Genome field, shedding light on opportunities for structure-based drug design based on established anti-viral targets found in our human genome. This work is the result of an outstanding collaboration between ROME’s exceptional structural biology team and world-class crystallographers.”