Scientists analyzed the chromosomes of three main animal species and discovered unexpected stability over more than 550 million years. Scientists have revealed that the genomes of marine invertebrates have remained remarkably stable over long periods of time. This new study, published in Science Advances, gives an overarching review of distantly related animal species, such as sponges, jellyfish, scallops, and the invertebrates most closely related to humans, and discovers that their chromosomes are remarkably similar.
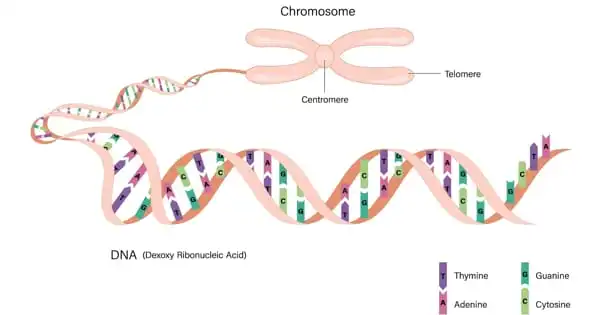
Consider the genome to be the instruction handbook for each cell, written in DNA code. It contains all of the hereditary information required for an organism’s operation. This instruction manual is organized into chapters (the chromosomes), which are further subdivided into pages (the genes).
“Because of random mutations, the arrangement of genes within chromosomes has gotten confused over deep time – at least 550 million years – similar to how pages within a chapter of a book get mixed up. More dramatically, we occasionally discover that two chromosomes have combined and become jumbled as if the chapters had been merged and scrambled.” Prof. Daniel Rokhsar, the paper’s last author and the principal investigator of the Molecular Genetics Unit at Japan’s Okinawa Institute of Science and Technology Graduate University (OIST), elaborated.
We see that genes might reside on the same chromosome in various species, but in distinct orders. In rare circumstances where two chromosomes fuse together and then get mixed up by scrambling across the newly fused chromosome, the fusion is irreversible and acts as a permanent marker of the chromosome’s evolutionary history.
Prof. Daniel Rokhsar
“However, we discovered a tremendous amount of stability overall. Despite the fact that the last common ancestor of these three groups lived more than half a billion years ago, many of their chromosomes are recognizable in the sense that they contain the same groupings of genes.”
The genomes of three broad groups were compared in the study: sponges (extremely primitive organisms with no muscles or nerves), cnidarians (particularly jellyfish and hydra), and bilaterians (scallops and amphioxus). These genomes had either previously been sequenced or were reported for the first time in this study. Despite the fact that several of these creatures had “draft” copies of their genomes sequenced previously, this early research fell short of allowing researchers to investigate general chromosomal arrangements.
Researchers can now put the puzzle pieces together and compare how genes are structured into lengthy threads thanks to advances in genetic technology. In this study, the hydra’s chromosomes were reconstructed for the first time, the amphioxus’ chromosomes were substantially improved, and a comprehensive comparative analysis was done.
The international team of researchers, which included scientists from OIST, the University of Vienna, the University of California campuses in Berkeley, Irvine, and Santa Cruz, the Ludwig Maximilian University of Munich, and the University College, London, discovered striking similarities between the chromosomes of the five different animals and confirmed that these similarities were also present in other animal genomes. In certain situations, they discovered chromosomal fusion patterns that were unique to certain animal subspecies. The researchers discovered four ancient fusions shared by scallops and several other mollusks, which mirrored a fusion in a draft genome of a marine worm.
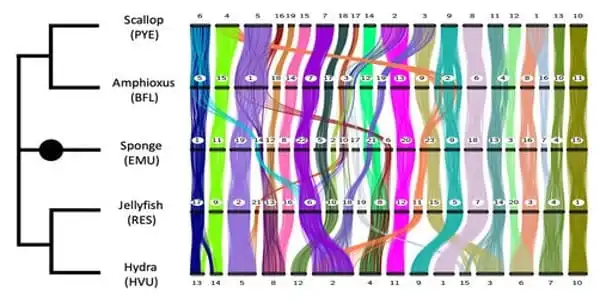
“We see that genes might reside on the same chromosome in various species, but in distinct orders,” Prof. Rokhsar noted. “In rare circumstances where two chromosomes fuse together and then get mixed up by scrambling across the newly fused chromosome, the fusion is irreversible and acts as a permanent marker of the chromosome’s evolutionary history. It’s like combining two decks of cards. We can keep rearranging them, but they will never again separate into two identical packs.”
When live creatures share the same fusions, the researchers conclude that the fusions occurred in an ancient common ancestor of these species. They have now made several testable predictions regarding genomes that have yet to be sequenced. For example, the researchers expect that the genomes of all mollusks and related “spiralian” creatures will have the same set of fusions found in scallops.
These findings highlight an intriguing conundrum. “Mammals have only been around for around 100 million years,” explained Prof. Rokhsar. “When we examine the genomes of two mammals, say a human and a mouse, the chromosomes appear to have been broken up into hundreds of pieces and then mingled together. The chromosome-scale conservation trait discovered in invertebrates is just not found in mammals.” He hypothesized that mammalian chromosomes evolved differently because mammals have historically lived in smaller groups than other marine invertebrates. Small groups allow these random mutations to survive, which may explain why chromosomal rearrangements propagate more easily in mammals.
The researchers discovered 29 ancestral chromosomal fragments in all. Furthermore, the researchers discovered that some of these segments existed 800-900 million years ago, before the formation of animals, when all organisms were unicellular or very basic multicellular. Thus, some genes have been traveling together for nearly a billion years, but the outcome of these ancient gene couplings is unknown.