Organelles in maintaining homeostasis, controlling development and aging, and producing energy, the bits and pieces of RNA and protein that make up a cell play critical roles in both health and sickness in humans.
Cells can differ in their organelles both between and within different cell types. Researchers can develop better therapies to cure a variety of disorders by better understanding cell function through the study of these distinctions.
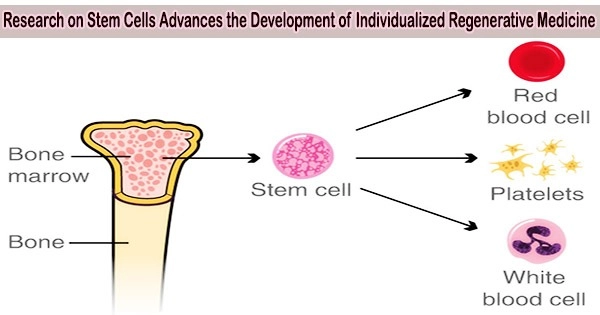
In two papers out of the lab of Ahmet F. Coskun, a Bernie Marcus Early Career professor in the Coulter Department of Biomedical Engineering at the Georgia Institute of Technology and Emory University, researchers examined a specific type of stem cell with an intracellular toolkit to determine which cells are most likely to create effective cell therapies.
“We are studying the placement of organelles within cells and how they communicate to help better treat disease,” said Coskun. “Our recent work proposes the use of an intracellular toolkit to map organelle bio-geography in stem cells that could lead to more precise therapies.”
Usually, the stem cells are used to repair defective cells or treat immune diseases, but our micro-study of these specific cells showed just how different they can be from one another. This proved that patient treatment population and customized isolation of the stem cells identities and their bioenergetic organelle function should be considered when selecting the tissue source. In other words, in treating a specific disease, it might be better to harvest the same type of cell from different locations depending on the patient’s needs.
Ahmet F. Coskun
Creating the subcellular omics toolkit
The first study examined mesenchymal stem cells (MSCs), which have historically presented intriguing therapeutic options for treating patients’ immune responses or mending damaged cells.
Through rapid subcellular proteomic imaging and a series of tests, the researchers were able to develop a data-driven, single-cell strategy that allowed for tailored stem cell treatments.
The scientists then utilized antibodies made to target particular organelles in a quick multiplexed immunofluorescence approach. They created maps by fluorescing antibodies and tracking wavelengths and signals to assemble images of numerous distinct cells.
The spatial arrangement of organelle interactions and the geographic distribution of related cells were then visualized on these maps, allowing researchers to predict which cell types would be most effective at treating specific disorders.
“Usually, the stem cells are used to repair defective cells or treat immune diseases, but our micro-study of these specific cells showed just how different they can be from one another,” said Coskun. “This proved that patient treatment population and customized isolation of the stem cells identities and their bioenergetic organelle function should be considered when selecting the tissue source. In other words, in treating a specific disease, it might be better to harvest the same type of cell from different locations depending on the patient’s needs.”
RNA-RNA proximity matters
The researchers expanded on their toolset in the following work, which was just published in Cell Reports Methods. They looked at the spatial arrangement of several nearby RNA molecules in a single cell, which is crucial to cellular function.
The technique was developed by the researchers by fusing spatial transcriptomics and machine learning. They discovered that studying the changes in gene proximity for cell type classification was more accurate than just studying gene expression.
“The physical interactions between molecules create life; therefore, the physical locations and proximity of these molecules play important roles,” said Coskun. “We created an intracellular toolkit of subcellular gene neighborhood networks in each cell’s different geographical parts to take a closer look at this.”
The project included two components: experiments at the lab bench and the creation of computational algorithms. To categorize RNA molecules according to their spatial distribution, the researchers looked at available datasets using an algorithm. This “nearest neighbor” algorithm helped determine gene groupings.
Then, to make it simple to find RNA molecules in individual cells, researchers labeled them with fluorescents on the bench. The distribution of RNA molecules revealed numerous characteristics, including the likelihood that genes are found in comparable subcellular regions.
If there are subtypes of unidentified cells in therapeutic cells, researchers cannot anticipate how these cells will behave when injected into patients. Cell therapy requires a large number of cells with highly identical characteristics.
More cells of the same type can be recognized using these techniques, and unique stem cell subsets with unusual gene repertoires can be extracted.
“We are expanding the toolkit for the subcellular spatial organization of molecules a ‘Swiss Army Knife’ for the subcellular spatial omics field, if you will,” said Coskun. “The goal is to measure, quantify, and model multiple independent but also interrelated molecular events in each cell with multiple functionalities. The end purpose is to define a cell’s function that can achieve high energy, Lego-like modular gene neighborhood networks and diverse cellular decisions.”