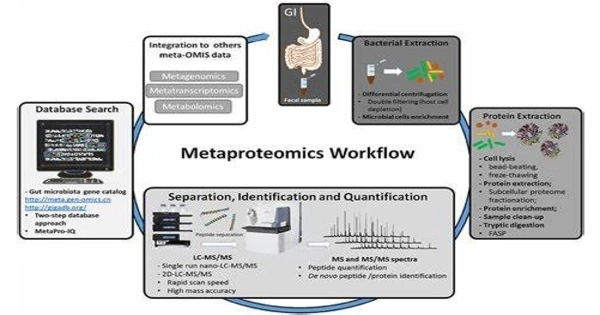
Metaproteomics is a branch of microbiology and environmental science that studies the collective set of proteins expressed by a complex microbial population. It is a catch-all word for all experimental ways to study all proteins in microbial communities and microbiomes derived from environmental sources. It gives information about the functional activities and metabolic processes of microorganisms in a certain ecosystem, such as a soil sample, a marine environment, or the human gut microbiome.
The term “metaproteomics” refers to the classification of investigations that deal with all proteins discovered and measured from complex microbial communities. Approaches to metaproteomics are similar to gene-centric environmental genomics or metagenomics. It is closely related to metagenomics, which is the study of these microbial communities’ genetic material (DNA).
Origin of the term
Francisco Rodrguez-Valera used the term “metaproteomics” to characterize the genes and/or proteins that are most abundantly expressed in environmental samples. The word “metagenome” was coined. Wilmes and Bond coined the term “metaproteomics” to describe the large-scale characterization of an ambient microbiota’s whole protein complement at a given point in time. Simultaneously, the words “microbial community proteomics” and “microbial community proteogenomics” are frequently used interchangeably to refer to various types of investigations and outcomes.
Here are some key points and concepts related to metaproteomics:
- Sample Collection: This usually starts with collecting environmental samples containing varied microbial groups. These samples can be obtained from a variety of sources, including soil, water, sediment, and biological samples such as excrement.
- Protein Extraction: The proteins are isolated from the microbial populations after the samples are collected. The microbial cells are broken down in this process to liberate their protein content.
- Protein Identification: After that, the extracted proteins are submitted to mass spectrometry, a powerful analytical technique that detects and quantifies the proteins in the sample. This data is used to generate a protein profile, often known as a proteome.
- Functional Analysis: Once the proteins are identified, researchers can gain insights into the metabolic processes and functions of the microbial community. This information can help in understanding the roles of different microbes within the ecosystem.
- Environmental and Medical Applications: It has a wide range of applications, from studying soil ecosystems to understanding the gut microbiome. It can be used in environmental monitoring, microbial ecology, and in the study of the human microbiome to better understand its impact on health.
Challenges
The diversity of microbial communities, the dynamic nature of proteomes, and the necessity for specialized analytical methods all contribute to the complexity of metaproteomics. Analyzing metaproteomic data can be computationally difficult and frequently necessitates the use of bioinformatics knowledge.
Metaproteomics has enhanced our understanding of the functional functions of microbial communities in many environments, and it has practical implications in sectors such as environmental science, biotechnology, and personalized medicine, where the human microbiome is important in health and disease.