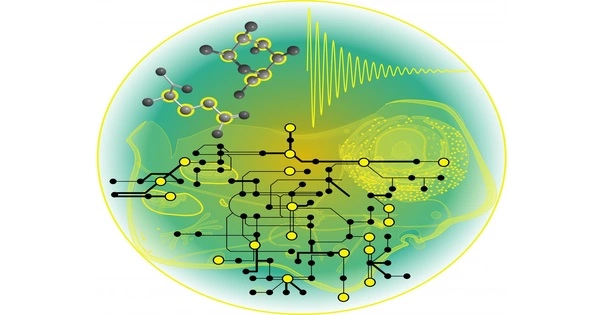
A metabolic network is an interconnected network of metabolic pathways found within a biological organism. It refers to the entire set of metabolic and physical processes that determine a cell’s physiological and biochemical properties. As such, these networks include metabolic chemical reactions, metabolic pathways, and the regulatory interactions that govern these reactions.
Metabolism is the sum of all chemical reactions that take place within a cell or organism in order to sustain life. Through various biochemical pathways, these reactions involve the conversion of molecules (substrates) into other molecules (products).
The sequencing of entire genomes has made it possible to reconstruct the network of biochemical reactions in a wide range of organisms, from bacteria to humans. Several of these networks are accessible via the internet, including the Kyoto Encyclopedia of Genes and Genomes (KEGG), EcoCyc, BioCyc, and metaTIGER. Metabolic networks are effective tools for researching and modeling metabolism.
Key features of metabolic networks include:
- Metabolic Pathways: These are biochemical reaction sequences that result in the synthesis or breakdown of a specific compound. Glycolysis, the citric acid cycle, and the pentose phosphate pathway are a few examples.
- Enzymes: Proteins that catalyze specific reactions and are essential components of metabolic pathways. Enzymes help to convert substrates into products by speeding up reactions that would otherwise be too slow to sustain life.
- Metabolites: Small molecules that participate in metabolic reactions. Sugars, amino acids, fatty acids, and other intermediates involved in cellular processes are examples of these.
- Regulation: Metabolic networks are tightly controlled to ensure that the appropriate reactions occur at the appropriate time and in the appropriate amounts. This regulation includes feedback loops, allosteric regulation, and enzyme activity control.
- Compartmentalization: In eukaryotic cells, metabolic reactions often occur in specific cellular compartments, such as the cytoplasm, mitochondria, or endoplasmic reticulum.
Uses
Comorbidity patterns in diseased patients can be detected using metabolic networks. Certain diseases, such as obesity and diabetes, can coexist in the same person, with one disease serving as a significant risk factor for the other. Disease phenotypes are typically the result of a cell’s inability to breakdown or produce an essential substrate.
An enzyme defect in one reaction, on the other hand, may affect the fluxes of subsequent reactions. These cascading effects link metabolic diseases with subsequent reactions, resulting in comorbidity. As a result, metabolic disease networks can be used to determine whether two disorders are linked due to correlated reactions.