In the past, researchers have achieved successful maturation of primate oocytes in the laboratory, including non-human primates like monkeys. These achievements have provided valuable insights into primate reproductive biology and have potential implications for human fertility research.
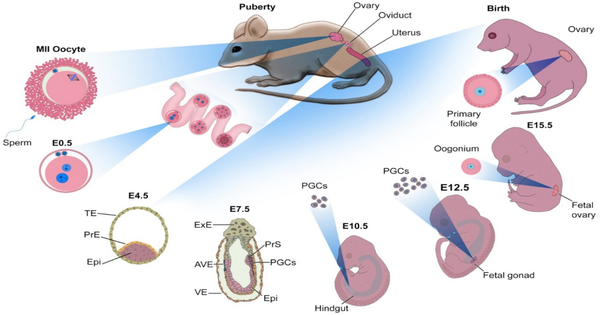
The many types of cells in the human body are produced through the process of differentiation, in which stem cells are converted to more specialized types. Currently, it is challenging for researchers to control the differentiation of stem cells in the lab (in vitro). Of particular interest are oocytes, which are female germ cells that develop into eggs. Understanding their development could have far-ranging impacts, from infertility treatment to conservation of endangered species.
A new study led by Dr. Mitinori Saitou of Japan has successfully induced meiotic (dividing) oocytes from embryonic stem cells of cynomolgus monkeys, which share many physiological traits with humans. The researchers hoped to shed light on germ cell development in humans and other primates by developing a culture method for inducing meiotic oocyte differentiation. The results of the study were published in the issue of The EMBO Journal.
We hope that our culture system can aid in the conservation of endangered species and the development of in vitro oocyte induction systems for other mammalian species with long lifespans.
Dr. Sayuri Gyobu-Motani
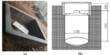
The researchers previously described conditions for inducing oogonia, or oocyte precursors, by aggregating human primordial germ cell-like cells (hPGCLCs) with cells from female mouse embryo ovaries and then culturing them under air-liquid interface conditions1. Similarly, cynomolgus monkey PGCLCs were stimulated to differentiate into oogonia but did not progress to meiotic oocytes. To overcome this barrier, the induced oogonia were isolated, re-aggregated with somatic cells from female mouse embryo ovaries, and cultured again.
The cynomolgus monkey oogonia were successfully induced to differentiate into meiotic oocytes under these new culture conditions, but their development was halted at the second stage of meiosis. The transcriptomic dynamics of oocytes in vitro (in the lab) were similar to those of oocytes in vivo (in our bodies), according to single-cell transcriptome analysis. The researchers also discovered differences in gene expression between in vitro and in vivo oocytes, implying a bottleneck for in vitro oocyte development that could lead to meiosis arrest in vitro.

Furthermore, the authors discovered that induced oocytes were involved in the genome-wide demethylation process in vitro, as seen in mouse and human female germ cell development, by performing whole-genome methylome analysis. They also discovered that demethylation behaved differently in X chromosomes derived from fathers and mothers. These distinct methylation dynamics were also discovered in human oogonia induced in vitro, implying that the mechanisms underlying female germ cell development may be shared by primates. As a result, this culture system could be useful as a model of primate germ cell differentiation.
When asked about the study’s potential impact, the authors stated that their method of re-creating multiple steps in the development of female germ cells may help to clarify the molecular mechanisms of primate oocyte development and may one day contribute to the treatment of impaired oocyte development in reproductive medicine.
“We hope that our culture system can aid in the conservation of endangered species and the development of in vitro oocyte induction systems for other mammalian species with long lifespans,” says first author Dr. Sayuri Gyobu-Motani.