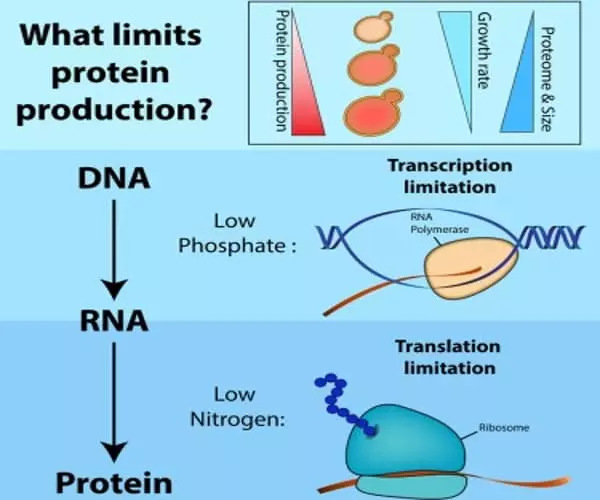
The genetic material of all eukaryotic organisms is stored in the cell nucleus in the form of DNA. To be used, this DNA must first be transcribed into messenger RNA in the cell cytoplasm, then translated into protein by ribosomes, which are small machines capable of decoding messenger RNA and synthesizing the appropriate proteins. However, the rate at which this mechanism occurs is not uniform: it must adapt in order for the protein to adopt the correct configuration. Deregulation of the production rate, in fact, causes structural flaws. Proteins that are not correctly folded aggregate, become unusable and are frequently toxic to the cell.
A team from the University of Geneva (UNIGE), Switzerland, in collaboration with the University of Hamburg, has demonstrated that the rate of protein synthesis is modulated by regulatory factors that change the rate of translation of messenger RNA into proteins at will by analyzing the rate of ribosome movement in yeast cells. These findings have been published in the journal Cell Reports.
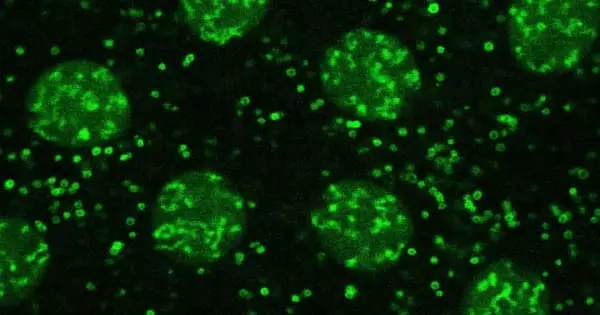
Proteins are three-dimensional structures that must interlock or interact with one another in order to function. When there is a structural flaw in a protein, it clumps together, becoming toxic and potentially pathological. This phenomenon has been observed in a variety of neurodegenerative diseases, including Alzheimer’s disease and amyotrophic lateral sclerosis. “We already knew that the rate at which proteins are made varies depending on need: sometimes fast, sometimes very slow,” explains Martine Collart, the study’s lead author and professor in the Department of Microbiology and Molecular Medicine at the UNIGE Faculty of Medicine. “However, we didn’t know how this mechanism was controlled at the time.”
We already knew that the rate at which proteins are made varies depending on need: sometimes fast, sometimes very slow. However, we didn’t know how this mechanism was controlled at the time. This methodology allows us to determine the position of ribosomes at any given time in the cell.
Martine Collart
Once an mRNA has been produced, the information contained in its nucleotide sequence is used to synthesize a protein via transcription and processing. Because DNA and RNA are chemically and structurally similar, the DNA can act as a direct template for the synthesis of RNA via complementary base-pairing. As the term transcription implies, it is as if a handwritten message is being converted, say, into a typewritten text. The language itself and the form of the message do not change, and the symbols used are closely related.
Ribosome profiling
To better understand this process, the researchers employed a novel and little-known technique known as ribosome profiling. “This methodology allows us to determine the position of ribosomes at any given time in the cell,” explains Olesya Panasenko, a researcher in Martine Collart’s laboratory and the head of the Faculty of Medicine’s ‘BioCode: RNA to Proteins’ Core Facility, who specialized in this technique. “It entails degrading all RNA that is not protected by the ribosome at a specific point in order to retain only the ribosome-protected fragments (RPFs). We then sequence these RPFs to determine how many ribosomes were on the mRNA and where they were located at the time. This reflects the translation’s speed and efficiency.”
The researchers looked at the speed and dynamics of protein production in both natural yeast cells and genetically modified yeast cells to see if there were any differences based on the genetic code. Small condensates of RNA and proteins appear in the cell during synthesis, slowing the rate of ribosome production. “The formation of these condensates is dependent on the presence or absence of regulatory factors known as Not, which act as decelerators,” Martine Collart explains. In the absence of these factors, the mechanism accelerates in the wrong places, resulting in aggregated proteins.
A speed regulated by the genetic code
Thus, Not factors bind to the ribosome at specific points during protein synthesis in order to slow down the ribosome during translation by condensing the RNA and the nascent protein. “One might wonder whether this regulatory mechanism is influenced by neurodegenerative diseases or by aging,” the authors speculate. As a result, it is possible that small disturbances, when added together, will have a significant cumulative effect over time.