Circadian rhythms are biological processes that have a roughly 24-hour period and are controlled by internal biological clocks. While circadian rhythms are well-researched in many organisms, including animals and plants, understanding bacterial circadian rhythms is a comparatively new development.
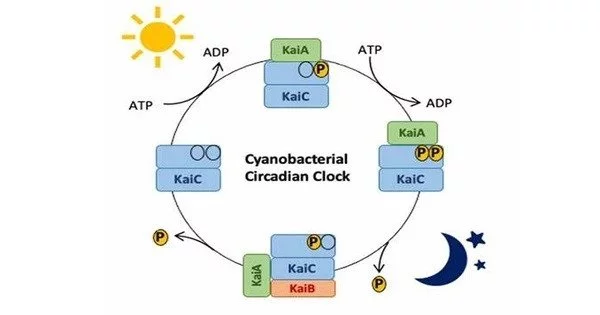
The best-studied example of bacterial circadian rhythms comes from the cyanobacterium Synechococcus elongatus (a type of photosynthetic bacteria). These bacteria contain an internal circadian clock that regulates several physiological activities, including photosynthesis timing. Synechococcus’ circadian clock is powered by a protein-based process.
Bacterial circadian rhythms, like all other circadian rhythms, are endogenous “biological clocks” with the three features listed below: In constant conditions (i.e. constant temperature and either constant light LL or constant darkness DD), they oscillate with a period that is close to, but not exactly, 24 hours long, (b) this “free-running” rhythm is temperature compensated, and (c) the rhythm will entrain to an appropriate environmental cycle.
Here’s a brief overview of the key components involved in the circadian clock of Synechococcus elongatus:
- KaiA, KaiB, and KaiC Proteins: These are the core components of the circadian clock. KaiC is a central player, and its phosphorylation state changes in a rhythmic manner over a 24-hour period. The KaiA and KaiB proteins interact with KaiC to modulate its phosphorylation.
- Phosphorylation and Dephosphorylation: The KaiC protein undergoes a rhythmic cycle of phosphorylation and dephosphorylation. This cycle is crucial for the proper functioning of the circadian clock.
- Output Pathways: Through output channels, the circadian clock governs a variety of cellular functions, including gene expression. These pathways involve clock components interacting with other cellular proteins.
- Regulation of Gene Expression: The circadian clock impacts gene expression timing in Synechococcus elongatus. Photosynthesis, nitrogen fixation, and other biological functions are all represented by genes in this category.
Until the mid-1980s, it was considered that only eukaryotic cells had circadian rhythms. Cyanobacteria (a phylum of photosynthetic eubacteria) currently exhibit well-documented circadian rhythms that match all of the criteria for true circadian rhythms. Three essential proteins whose structures have been discovered in these bacteria can form a molecular clockwork that orchestrates global gene expression. This mechanism improves the fitness of cyanobacteria in rhythmic settings.
The study of bacterial circadian rhythms is not confined to Synechococcus elongatus. Other bacteria, including some non-photosynthetic species, exhibit circadian-like activities. Understanding bacterial circadian rhythms has significance for domains such as synthetic biology, as researchers investigate ways to alter bacteria’s circadian clocks for various applications.