A chemical and biomolecular engineer with biosecurity expertise in teaching cells to create and use chemical building blocks that do not exist in nature. New research describes advancements in the stability of a biocontainment strategy that relies on a microbe’s reliance on a synthetic nutrient to keep it contained. Scientists and engineers are always looking for new ways to improve our world.
Synthetic biology is a new field that has the potential to improve our ability to manufacture chemicals, develop therapeutic medicines such as biopharmaceuticals and vaccines, and improve agricultural production, among other things. It works by combining natural or engineered DNA fragments in new ways in biological systems such as microbes, bacteria, or other organisms.
According to Aditya Kunjapur, assistant professor of chemical and biomolecular engineering at the University of Delaware, as these sophisticated microbial technologies advance, scientists must investigate ways to keep these organisms from ending up in the wrong environment.
A bacterium that is good at producing large amounts of chemicals, for example, is great in a bioreactor but isn’t necessarily what we want in the environment or in our food. Similarly, while a specialized bacterium developed to aid plant productivity is beneficial in agriculture, we may not want that same bacterium to end up in our bodies.
You can take enzymes that bacteria need to grow and make them rely on a man-made building block that they will never see in nature. We discovered that the DEP E. coli was unable to “escape” or grow outside the cell in three parallel experiments.
Aditya Kunjapur
One promising solution is to engineer traits directly into the microbe to create built-in safeguards to control where it can grow, a process known as biological containment. “Being able to control where microbes grow will aid in the development of safer technologies to treat disease and clean up the environment,” said Kunjapur, an emerging leader in biosecurity with expertise in teaching cells to create and harness chemical building blocks not found in nature.
Kunjapur is now the lead author of a new paper published in Science Advances that describes progress on the stability of a biocontainment strategy that was first reported in 2015 and uses a microbe’s dependence on a synthetic nutrient to keep it contained. The research was carried out in collaboration with Harvard Medical School (HMS), and co-authors are now employed at HMS, UD, Johns Hopkins University, and industry.
Under lock and key
In the paper, the researchers looked at a strain of Escherichia coli (E. coli) that had been engineered to rely on a synthetic nutrient for survival, a process known as synthetic auxotrophy. They discovered that after growing the strain for 100 days, it remained stable and retained its dependence.
“You can take enzymes that bacteria need to grow and make them rely on a man-made building block that they will never see in nature,” Kunjapur explained.
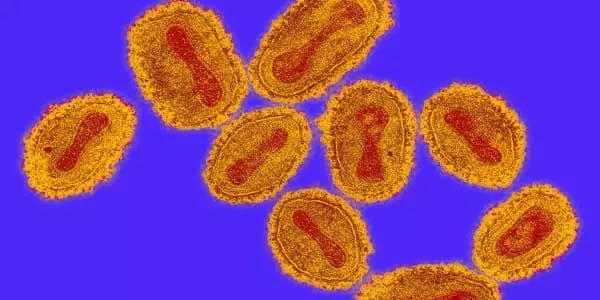
The research builds on previous work done in the Church lab at Harvard Medical School, where the majority of the research was done and Kunjapur did postdoctoral work. The researchers demonstrated in that study that if one enzyme required this synthetic building block, bacteria would quickly find a way out. However, because three enzymes required the synthetic building block, the bacteria couldn’t “unlock the cell door” within a 24-hour period to flee.
“Asking the microbe to mutate its genome in precisely three different ways at the same time is asking too much, and it will not find that solution,” Kunjapur explained. “Thus, while the same ‘key’ may open all three doors, the microbe cannot be present at all three doors at the same time to open them, so it remains contained.”
The question was whether, given more time to mature, they would find a way out. In this new study, the researchers extended the study and grew the E. coli strain, dubbed DEP, continuously for 100 days while also applying extra stress by gradually decreasing the amount of synthetic nutrient the bacteria received.
The researchers discovered that the DEP E. coli was unable to “escape” or grow outside the cell in three parallel experiments. It did, however, adapt to a lower concentration of the synthetic nutrient it required to survive (ten times less). Furthermore, the researchers discovered that proteins on the surface of the host cell changed, which had never been observed before.
“We believe that as the amount of nutrients was reduced, the DEP E. coli modified the proteins on the cell’s surface that regulate how much synthetic nutrient is allowed inside in order to retain more nutrients inside and keep growing,” Kunjapur explained. “We were surprised because we didn’t expect them to adapt in the first place, and then we didn’t expect them to adapt in this way.”
To validate their findings, the researchers engineered the same genetic traits into the ancestor E. coli strain and discovered it could grow at lower synthetic nutrient concentrations from the start. According to Kunjapur, this information could be useful for scaling the experiment in the future.
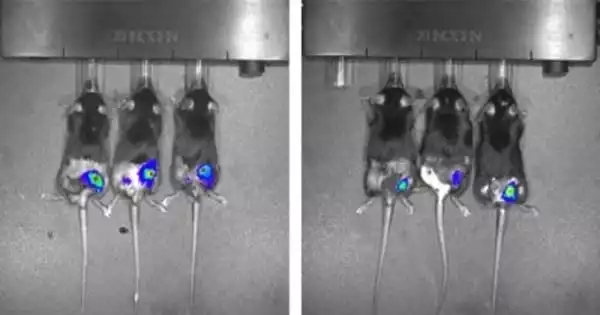
Kunjapur’s colleague Eriona Hysolli, a postdoctoral fellow in the Church lab, performed a follow-up experiment in which the bacteria were directly added to mammalian cells. The outcomes were intriguing. While the bacteria grew rapidly in the first few days, they simply faded away over the course of a week, without the need for antibiotics.
“Our findings are the first to demonstrate the use of recoded bacterial strains with controllable growth in a mammalian cellular model. We are excited about the gut microbiome’s promising applications in human health “said Hysolli, a paper co-author.
Building on this work, Michaela Jones, a UD doctoral student in Kunjapur’s lab and paper co-author, carried out additional experiments that revealed the bacteria stopped growing when the synthetic nutrient was removed, then resumed growth when the nutrient was replaced. According to Kunjapur, these findings provide researchers with new tools for controlling how and when bacteria grow by regulating the nutrients they require to survive.
“Now, if you need bacteria to grow at a specific rate or time, we have a synthetic building block that can act as a lever or knob to control that,” he explained. Kunjapur intends to investigate the biocontained DEP E. coli strain, as well as other microbe designs, under a broader range of conditions in the future to see if the approach will hold up in the environment. He is also working on biomedical applications for live bacterial strains that could be used as therapeutics, probiotics, and vaccines.