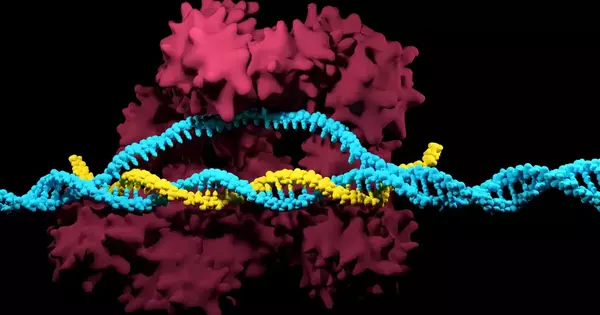
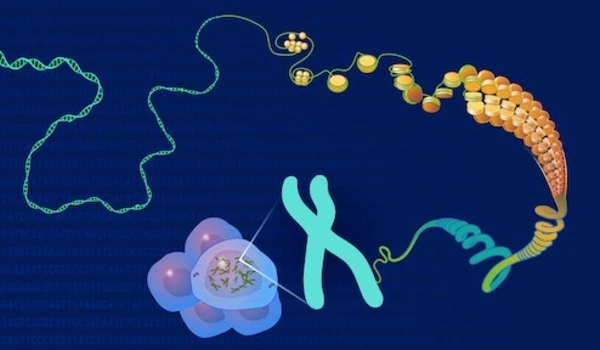
The genome is an organism’s complete set of genetic material (DNA). It contains all of the information required for an organism’s development, functioning, and reproduction. The human genome, for example, is made up of roughly 3 billion base pairs of DNA.
Plants exhibit a wide range of traits relevant to breeding, such as plant height, yield, and pest resistance. One of the most difficult challenges in modern plant research is determining the genetic differences that cause this variation.
A research team led by the “Crop Yield” working group at Heinrich Heine University Düsseldorf (HHU) and the Carnegie Institution of Science at Stanford has developed a method to precisely identify these special differences in genetic information. In the journal Genome Biology, they demonstrate the great potential of their method by using maize as an example, and present regions in the maize genome that may help to increase yields and pest resistance during breeding.
DNA contains the blueprint for all organisms. This includes the genes that encode proteins and determine the inherent characteristics of an organism. There are also other important sections of DNA, particularly those that control gene regulation, i.e. when, under what conditions, and to what extent genes are activated.
Unlike protein-coding genes, regulatory sites usually cannot be identified based on the sequence alone. This makes them very difficult to pinpoint. Our method uses hybrid plants to measure the direct effects of variation in DNA sequence on transcription factor binding.
Professor Dr. Zhi-Yong Wang
However, when compared to genes, these regulatory regions – also known as “cis elements” – are difficult to find. Changes in these DNA elements, however, are largely responsible for the differences between organisms – and thus between different plant varieties.
Researchers have discovered that regulatory regions are the binding sites for specific proteins over the last few decades. Transcription factors are responsible for determining when and for how long genes are activated.
Dr. Thomas Hartwig, co-corresponding author and leader of the Crop Yield research group at HHU’s Institute for Molecular Physiology and the Max Planck Institute for Plant Breeding Research (MPIPZ) in Cologne, says, “Finding the few variations that are key to changing traits such as pest resistance among the millions and millions of non-causative genome differences is the ultimate search for a needle in a haystack.”
“Unlike protein-coding genes, regulatory sites usually cannot be identified based on the sequence alone. This makes them very difficult to pinpoint. Our method uses hybrid plants to measure the direct effects of variation in DNA sequence on transcription factor binding,” says lead author Professor Dr. Zhi-Yong Wang from the Carnegie Institution for Science.
Researchers from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben, as well as the University of Nebraska-Lincoln and Iowa State University in the United States, collaborated on the study.
The research team can compare which regulatory regions differ across the entire genome by using hybrids, which are the first generation of plants created by crossbreeding two varieties. Dr. Julia Engelhorn, a co-author, says, “Our analytical method allows us to precisely measure whether transcription factors bind more to the maternal or paternal genome.” This method also allowed the team to identify thousands of differences associated with traits in maize, such as yield and pest resistance.
The technology was demonstrated for a transcription factor in the brassinosteroid pathway, a hormone related to growth and disease. Institute director Professor Dr. Wolf B. Frommer: “The team has identified thousands of genomic variations that can explain why one variety of maize behaves differently in terms of its yield or resistance to disease. Moreover, the team was able to show that these differences are almost equally genetic and epigenetic.” The latter describes processes that influence gene activity without being encoded in the DNA sequence itself.
The study’s main finding is a list of more than 6,000 genome regions that can be targeted for plant breeding. These could include areas where certain maize varieties express positive traits that other plants do not.
“Knowing where in the genome modern breeding methods can be used to transfer characteristics from one variety to another is critical for biotechnology,” says Hartwig. Our research could help others find these intriguing genome regions. The study findings lay the foundation for using modern techniques to cultivate new varieties of maize by skillfully combining the optimal variants,” Professor Frommer adds.