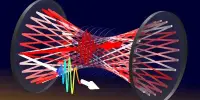
DNA nanotechnology is concerned with the design and construction of nanostructures utilizing DNA as a building material. It could be used in medicine, electronics, and other fields. An multinational team of scientists have discovered a revolutionary form of DNA nano engine. It is propelled by a smart mechanism and may make pulsing movements. The researchers intend to equip it with a coupling and use it as a motor in complicated nanodevices. Their findings were just published in the journal Nature Nanotechnology.
Petr Šulc, an assistant professor at Arizona State University’s School of Molecular Sciences and the Biodesign Center for Molecular Design and Biomimetics, has collaborated with Professor Famulok (project lead) from the University of Bonn, Germany, and Professor Walter from the University of Michigan on this project.
Šulc has used computer modeling tools from his group to obtain insight into the design and operation of this leaf-spring nano engine. The structure is made up of about 14,000 nucleotides, which are the fundamental structural components of DNA.
“Being able to simulate motion in such a large nanostructure would be impossible without oxDNA, the computer model that our group uses for design and design of DNA nanostructures,” explains Šulc. “This is the first successfully engineered chemically powered DNA nanotechnology motor. We are really happy that our research methods may be useful in researching it, and we hope to construct even more complicated nanodevices in the future.”
Being able to simulate motion in such a large nanostructure would be impossible without oxDNA, the computer model that our group uses for the design and design of DNA nanostructures. This is the first successful engineered chemically powered DNA nanotechnology motor. We are really happy that our research methods may be useful in researching it, and we hope to construct even more complicated nanodevices in the future.
Petr Šulc
This unique engine is similar to a hand grip strength trainer in that it strengthens your grip when used on a regular basis. The motor, on the other hand, is one million times smaller. A spring connects two handles in a V-shaped construction.
Squeeze the handles together against the spring resistance in a hand grip strength trainer. When you let go of the handles, the spring returns them to their original position. “Our motor operates on a very similar principle,” says Professor Michael Famulok of the University of Bonn’s Life and Medical Sciences (LIMES) Institute. “But the handles are not pressed together but rather pulled together.”
The researchers have repurposed a mechanism without which there would be no plants or animals on Earth. Every cell is equipped with a sort of library. It contains the blueprints for all types of proteins that each cell needs to perform its function. If the cell wants to produce a certain type of protein, it orders a copy from the respective blueprint. This transcript is produced by the enzymes called RNA polymerases.
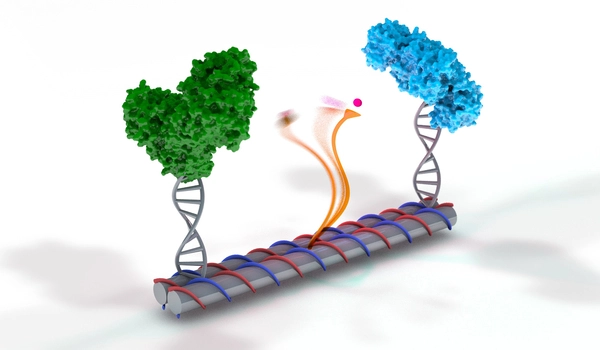
RNA polymerases drive the pulsing movements
Long strands of DNA comprise the original blueprint. The RNA polymerases proceed along these strands, letter by letter, copying the stored information. “We took an RNA polymerase and attached it to one of the handles in our nanomachine,” says Famulok, who is also a member of the transdisciplinary research areas “Life & Health” and “Matter” at the University of Bonn.
“We also strained a DNA strand between the two handles in close proximity.” This strand is grabbed by the polymerase in order to be copied. It drags itself along the strand, making the non-transcribed region smaller and smaller. This gradually pushes the second handle towards the first, compressing the spring at the same time.”
The DNA strand between the handles contains a particular sequence of letters shortly before its end. This so-called termination sequence signals to the polymerase that it should let go of the DNA. The spring can now relax again and moves the handles apart. This brings the start sequence of the strand close to the polymerase and the molecular copier can start a new transcription process: The cycle then repeats. “In this way, our nanomotor performs a pulsing action,” explains Mathias Centola from the research group headed by professor Famulok, who carried out a large proportion of the experiments.
An alphabet soup serves as fuel
This motor, like any other form of motor, requires energy. It comes from the “alphabet soup” from which the polymerase generates the transcripts. Each of these letters (technically, nucleotides) has a short tail made up of three phosphate groups – a triphosphate. The polymerase must remove two of these phosphate groups in order to attach a new letter to an old phrase. This releases energy, which it can then use to connect the letters. “Our motor thus uses nucleotide triphosphates as fuel,” according to Famulok. “It can only continue to run when a sufficient number of them are available.”
The researchers proved that the motor may be simply integrated with different architectures. This should allow it to move across a surface, much like an inchworm does when it pushes itself along a branch in its own unique manner. “We are also planning to produce a type of clutch that will allow us to only utilize the power of the motor at certain times and otherwise leave it to idle,” said Famulok. In the long run, the motor might become the brain of a sophisticated nanomachine. “However, there is still a lot of work to be done before we reach this stage.”
Šulc’s lab is highly interdisciplinary and applies broadly the methods of statistical physics and computational modeling to problems in chemistry, biology and nanotechnology. The group develops new multiscale models to study interactions between biomolecules, particularly in the context of design and simulations of DNA and RNA nanostructures and devices.
“Just as complex machines in our everyday use — planes, cars and chips in electronics — require sophisticated computer-aided design tools to make sure they perform a desired function, there is a pressing need to have access to such methods in the molecular sciences.”
Professor Tijana Rajh, director of the School of Molecular Sciences, said, “Petr Šulc and his group are doing extremely innovative molecular science, using the methods of computational chemistry and physics to study DNA and RNA molecules in the context of biology as well as nanotechnology. Our younger faculty members in the School of Molecular Sciences have an extraordinary record of achievement, and Professor Šulc is an exemplar in this regard.
Bio-nanotechnology
The basic molecules of life are DNA and RNA. In live cells, they perform a variety of roles, including information storage and transport. They have also shown promise in nanotechnology, where customized DNA and RNA strands are utilized to create tiny structures and gadgets. In his own words, “It is a little bit like playing with Lego blocks except that each Lego block is only a few nanometers (a millionth of a millimeter) in size, and instead of putting each block into the place where it should go, you put them inside a box and shake it randomly until only the desired structure comes out.”
This process is called self-assembly, and Šulc and his colleagues use computational modeling and design software to come up with the building blocks that reliably assemble into the shape one wants at nanoscale resolution.