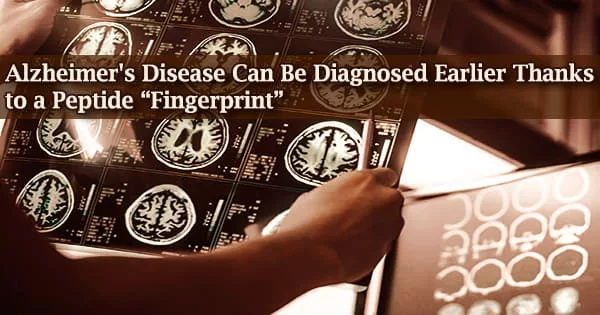
When proteins or peptides fold incorrectly (misfold), their spatial structure is altered, leading to neurodegenerative disorders including Alzheimer’s and Parkinson’s.
This is the outcome of slight variations in the biomolecules’ chemical make-up. A quick and efficient technique has been devised by scientists at the Karlsruhe Institute of Technology (KIT) to identify such misfolding at an early stage of the illness.
The structure of dried protein and peptide solution residue reveals misfolding. The technique uses neural networks to analyze micrographs and has a > 99% prediction accuracy. Advanced Materials has published the findings.
A degenerative neurologic condition called Alzheimer’s disease results in the death of brain cells and brain shrinkage. The most prevalent form of dementia, which is characterized by a steady decline in thinking, behavioral, and social abilities and impairs a person’s capacity for independent functioning, is Alzheimer’s disease.
Multiple brain functions are impacted. In most cases, mild memory issues are the initial indication of Alzheimer’s disease. The prevalence of Alzheimer’s disease is highest among those over 65. Age-related increases in the chance of developing Alzheimer’s disease and other forms of dementia are shown in 1 in 14 people over the age of 65 and 1 in 6 people over the age of 80.
Proteins and peptides’ biological functions are governed by their biochemical structures. There are several signs that even little alterations in space or structure might encourage the growth of illnesses.
Protein and peptide misfolding, which is brought on by such alterations, has been linked to a number of neurodegenerative disorders. A single amino acid residue separates each of the amyloid beta (A-42) peptides, which are hereditary mutations of Alzheimer’s disease and play a significant part in the disease.
Since the structures are very similar and difficult to distinguish with the naked eye, it was definitely a surprise that the neural networks were so effective. The stain patterns of amyloid beta peptides serve as exact fingerprints that reflect the structural and spatial identity of a peptide.
Professor Jörg Lahann
There hasn’t been a quick and reliable way for foretelling protein mutations up until now. A research team under the direction of Professor Jörg Lahann at KIT’s Institute of Functional Interfaces (IFG) has created a technique for misfolding detection using the structure of dried protein and peptide solutions.
“The stain patterns were not only characteristic and reproducible but also result in a classification of eight mutations with a predictive accuracy of more than 99 percent,” said Lahann, author of the study, in describing the results.
The team demonstrated that important details about the peptides’ fundamental and secondary structures may be deduced from the stains left behind when droplets of peptide solution dry on a solid surface.
Stain Patterns as Exact Peptide Fingerprints
An automated pipetting device accurately places the protein and peptide solutions on glass slides to achieve controlled and repeatable results. The slides’ surfaces were previously coated with a hydrophobic polymer.
The researchers used polarization microscopy to capture images in order to examine the intricate stain patterns from the dried droplets. After that, deep learning neural networks were used to examine the photos.
“Since the structures are very similar and difficult to distinguish with the naked eye, it was definitely a surprise that the neural networks were so effective,” says Lahann about the results. “The stain patterns of amyloid beta peptides serve as exact fingerprints that reflect the structural and spatial identity of a peptide.”
According to Lahann, this technology makes it possible to identify Alzheimer’s variations with the highest level of clarity in a matter of minutes.
Simple Sample Preparation Delivers Fast Diagnoses
The findings imply that a technique as straightforward as drying a droplet of peptide solution on a solid surface can act as a marker for minute variations in the main and secondary structures of peptides.
“Scalable and accurate detection methods for the stratification of conformational and structural protein alterations are urgently needed in order to decode the pathological signatures of diseases like Alzheimer’s and Parkinson’s,” says Lahann.
Additionally, it is a reasonably easy procedure that allows for a quick and patient-friendly diagnosis because it does not call for complicated sample preparation. The technique also holds considerable promise for other uses in disease molecular identification and medical diagnostics.