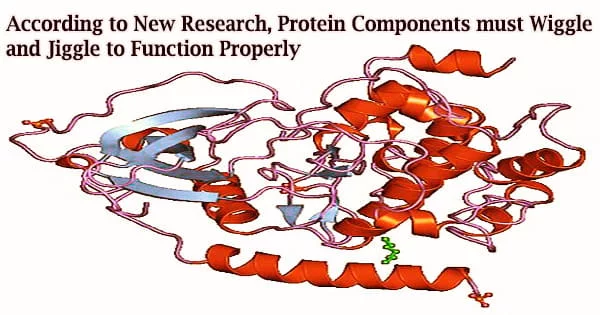
Researchers from Johns Hopkins Medicine say they have looked at the atomic structure of proteins to contribute to the growing body of evidence showing proteins’ trembles, trembles, and quivers are essential to their capacity to operate.
The results of the study could aid in the development of novel medications that can alter or disrupt the complex “dances” of proteins to change their functions. The July 15, 2022, issue of Science Advances will contain the findings of the researchers’ investigations.
Along with enzymes, which coordinate chemical changes within cells, proteins are organic substances with blueprints found in DNA. They serve as the “business ends” of biology, forming the structural elements of tissues.
Although scientists have contested the significance of this “dancing” act, Dominique Frueh, Ph.D., associate professor of biophysics and biophysical chemistry at the Johns Hopkins University School of Medicine, notes that proteins have long been known to wriggle and move.
“The way proteins engage with the right partner at the right time essentially, how they communicate is very important for understanding their function,” he says, “and we have found that protein wiggles are critical for this communication.”
Frueh’s team investigated the movement of the HMWP2 protein, a nonribosomal peptide synthetase, in an effort to advance this understanding. These enzymes’ several domains, or different areas, function as an assembly line to create complex natural products from simple molecules.
These organic ingredients frequently have medicinal characteristics, including bacitracin, which is used in topical antibiotic ointments. In the case of HMWP2, its end result is yersiniabactin, an iron-scavenging molecule for bacteria like Escherichia coli, which causes uTIs, and Yersinia pestis, which causes bubonic plague.
According to Frueh, by comprehending how protein domains interact, scientists may be able to alter the domains’ ability to synthesize new compounds.
The team discovered that the HMWP2 enzyme’s one domain can link with multiple partner domains simultaneously as a result of broad domain wriggling.
We found that the two domains would only bind to one another as the second domain gets modified, which means they would only engage as needed for making the product and avoid wasting time together when the second domain is not modified. Somehow, the first domain is able to sense when the second domain is modified, and we sought to investigate whether motions played a role in this recognition process.
Dominique Frueh
Nuclear magnetic resonance (NMR) spectroscopy, a technique that employs strong magnetic fields to examine the molecular environments of nuclei within the centers of atoms, was used to track the motion of one of HMWP2’s domains down to each individual atom in the molecule. This allowed the researchers to determine the significance of protein movement.
Although NMR is frequently used to establish the structures of tiny proteins, it is challenging to use the technology to follow the motions within large proteins. To address this issue, Frueh’s team, which also included NMR expert Subrata Mishra, Ph.D., graduate student Kenneth Marincin, and postdoctoral fellow Aswani Kancherla, Ph.D., used naturally occurring nitrogen-15 and carbon-13 forms of nitrogen and carbon to flag two domains of the HMWP2 enzyme and track the change in motion of one domain when a second domain was modified, as happens when the enzyme makes its natural product.
“We found that the two domains would only bind to one another as the second domain gets modified, which means they would only engage as needed for making the product and avoid wasting time together when the second domain is not modified,” says Frueh. “Somehow, the first domain is able to sense when the second domain is modified, and we sought to investigate whether motions played a role in this recognition process.”
Additionally, they discovered modifications in motion throughout the entire carbon-13c-flagged domain, including at a second, distant binding point that a third domain uses in addition to where it binds to the first domain.
These two spots on HMWP2 are 40 billionths of a meter apart on an atomic scale, according to Frueh. Furthermore, the scientists were particularly interested in how they connect despite being far apart.
The researchers genetically modified HMWP2 proteins with a mutation that happened in a place on the domain far from the two spots they had found in order to demonstrate that motions facilitated the interaction with the remote site and the sensing of the second domain modification. Therefore, the mutation did not completely prevent the sites from interacting with other domains.
“We found that the protein domain was structurally stable, but all of its movement was hindered,” says Frueh.
The researchers showed that the motions inside the protein were essential for the domains to cooperate and that the mutant protein’s lack of motion hindered its capacity to interact with other domains even when they were corrected.
According to Frueh, a precise understanding of how proteins move could be used by researchers to create brand-new medications that don’t target a protein’s natural active location but instead slow it down to render it inactive. According to him, such a strategy might give designers more freedom to create drugs with fewer undesirable side effects.
According to Frueh, scientists are investigating how computation and artificial intelligence might enhance our knowledge of and ability to forecast protein mobility.
Funding for the research was provided by the National Institutes of Health’s National Institute of General Medical Sciences and the National Institute of Allergy and Infectious Diseases.
Other researchers who contributed to the study are Guillaume Bouvignies from the École Normale Supérieure in Paris; Santrupti Nerli from the University of California, Santa Cruz; Nikolaos Sgourakis from the University of Pennsylvania and Daniel Dowling from the University of Massachusetts.