RNA sequencing, or RNA-seq, is a method used to analyze the transcriptome, or the set of all RNA molecules present in a cell or tissue. A “pantranscriptome” is a term used to describe the analysis of all RNA molecules, including those that do not code for proteins (such as non-coding RNAs) as well as those that do.
To understand how much a person’s genes are “turned on” and carry out functions in the body, it is necessary to map their RNA landscape to a common reference. However, when the reference does not offer sufficient details to for precise mapping, a problem known as reference bias might arise.
The first method for genome-wide analysis of RNA sequencing data using a “pantranscriptome,” which combines a transcriptome and a pangenome, a reference containing genetic material from a cohort of various individuals, as opposed to just a single linear strand, is presented by researchers at UC Santa Cruz in a new paper published in the journal Nature Methods.
A group of scientists led by UCSC Associate Professor of Biomolecular Engineering Benedict Paten have released a toolkit that allows researchers to map an individual’s RNA data to a much richer reference, addressing reference bias and leading to much more accurate mapping.
“This is pangenome plus transcriptome that combination has never really been done before until now,” said Jordan Eizenga, the paper’s co-first author and a postdoctoral scholar in the UCSC Computational Genomics Lab. “This is the first time anyone has attempted to incorporate the pangenome as a standard feature of the RNA sequencing mapping.”
Researchers attempting to understand gene expression through RNA sequencing analysis will find this tool helpful. The tools are publicly available and can be accessed via Github.
“With this toolkit, we are employing this more diverse data that we can now get from the pangenome to improve the measurement of gene expression data, something that can widely vary between individuals,” Paten said. “The aim is to make the impact of this more diverse data felt on studies that are looking at gene expression, resulting in better analysis for cell models, organoid models, and other research applications.”
Although scientists once believed that RNA’s only known use was in the translation of DNA into proteins, they now know that the vast majority of RNA is non-coding and does not produce proteins; instead, it can perform roles like affecting cell shape or controlling genes.
It’s definitely a very forward-looking study in that other genome-wide expression methods are not yet really utilizing pangenomes and haplotype information. We’re now thinking ahead as to what pangenomics might additionally bring to the table in transcriptomic analyses.
Jonas Sibbesen
Researchers can better comprehend a person’s gene expression by mapping the transcriptome, which is the term for the complete RNA landscape. The pantranscriptome advances the recently popularized term “pangenomics” in the field of genomics.
Scientists typically compare an individual’s genome to a reference genome made up of a single linear strand of DNA bases when assessing an individual’s genomic data for variance. Using a pangenome, researchers can simultaneously compare a person’s genome to a cohort of reference sequences that reflect a variety of biogeographic ancestries and are genetically diverse. This gives the researchers more data points to compare in order to comprehend a person’s genomic variance.
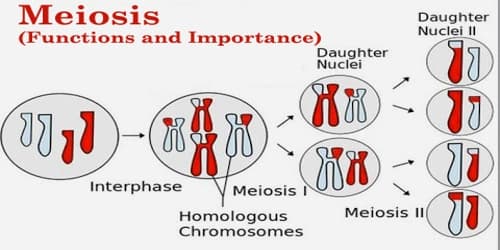
Because RNA sequences are spliced by physiological mechanisms, one set of RNA data may originate from disconnected regions of the genome, making it difficult to accurately match them to a reference, mapping RNA sequencing data to understand gene expression can be tricky. These splicing sites vary from person to person and are not constant across the human population.
Furthermore, it is challenging to determine if the collection of genes precisely originates from the set of chromosomes inherited from the person’s mother or the set received from the person’s father.
However, using a new pipeline of open source tools, scientists can now take the spliced segments of a person’s RNA, map where they align on a pangenome, determine which haplotype the data belongs to, and study gene expression.
The process first identifies the regions of the genome including the splice sites from which the RNA sequencing data originates and marks those locations on the pangenome reference. Then, those highlighted regions are contrasted with a pantranscriptome made up of transcripts specific to each haplotype created using the pangenome’s reference data. This process calls for sophisticated, difficult algorithmic techniques.
On the basis of this comparison between the mapped data and the transcripts in the pantranscriptome, it creates estimates of the levels of gene expression and determines which haplotypes the genes belong to.
“It’s definitely a very forward-looking study in that other genome-wide expression methods are not yet really utilizing pangenomes and haplotype information,” said Jonas Sibbesen, co-first author on the study and a former postdoctoral scholar in the UCSC Computational Genomics Lab who is now an assistant professor at the University of Copenhagen. “We’re now thinking ahead as to what pangenomics might additionally bring to the table in transcriptomic analyses.”
Future work by the researchers will focus on further improving these tools to make them helpful for downstream informatics analysis and customizing them for the unique requirements of single-cell data study. The team anticipates that their new toolset will demonstrate the value of pangenomics-derived analysis for the time being.
“We need to be able to explain to some researchers how a pangenome reference will benefit them,” Paten said. “This pipeline is really a first go at doing this for RNA, for functional data, for expression data.”