Plants frequently form communities with microbes in their roots, which affects plant health and development. Although a variety of factors influence microbe recruitment, it is unclear if genetic variation in host plants plays an effect. In a new study, researchers from the University of Illinois Urbana-Champaign investigated this question, and their findings can assist boost agricultural output.
“Previously, researchers have only looked at what kinds of microbes are present in association with plants, but not what might be driving the formation of these communities and how we might be able to control these drivers through plant breeding,” explained Angela Kent (CABBI), a professor of natural resources and environmental sciences.
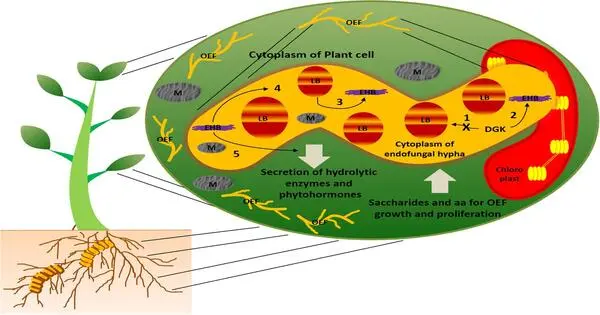
Microbes develop complex communities known as microbiomes in and around plants’ roots. Chemical cues allow the host plants to control which bacteria are welcomed into their roots, known as endophytes. They can also change the soil conditions around the roots, influencing which bacteria can live on the root surface, or rhizosphere. However, in order to breed plants based on the microorganisms they associate with, researchers must first understand how plant genomes affect the rhizosphere microbiome.
Crop breeding is based on yield. However, we must adopt a broader perspective and assess how bacteria can assist to crop sustainability. The attractiveness of dealing with wild plants is that there is a great deal of genetic variety to study.
Angela Kent
To answer this question, the researchers examined two native silver grass species, Miscanthus sinensis and Miscanthus floridulus. These plants are considered promising bioenergy crops since they require lower fertilizer concentrations to produce more growth than traditional crops.
The study was conducted in 16 sites across Taiwan and included a range of environmental conditions, such as hot springs, mountain peaks, and valleys, to represent all possible environmental extremes. The researchers collected 236 rhizosphere soil samples from randomly selected Miscanthus plants and also isolated the microbiome inside the roots.
“Although the scale of this study was unprecedented, we were mindful of the plant protection and quarantine regulations. We processed the samples in Taiwan to extract the endophytic microbial community and collect the rhizosphere microbiome,” Kent said.
The researchers employed two different DNA sequencing techniques in their investigation. The microbiomes in and around the roots were discovered by analyzing the DNA sequences of bacterial and fungal rRNA genes, with an emphasis on the region of the genome that is unique to each species. Microsatellites, little fragments of repetitive DNA that can differentiate even closely related plant populations, were used to quantify variation in the plant genome.
“The samples were collected 15 years ago, when the project was too large for the sequencing capabilities at the time. As the cost of sequencing came down, it allowed us to revisit the data and take a closer look at the microbiome. During sample processing, we also inadvertently extracted plant DNA and we were able to use that as a resource for genotyping our Miscanthus populations,” Kent said.
“We screened the host genome sequences for insights into how they can affect the microbiome,” said Niuniu Ji, a postdoctoral researcher in the Kent lab. “I discovered that the plants affect the core microbiome, which was exciting.”
Although plant microbiomes are very diverse, the core microbiome is a collection of microbes that are found in most samples of a particular set of plants. These microbes are considered to play an important role in organizing which other microbes are associated with the plant and helping with host growth.
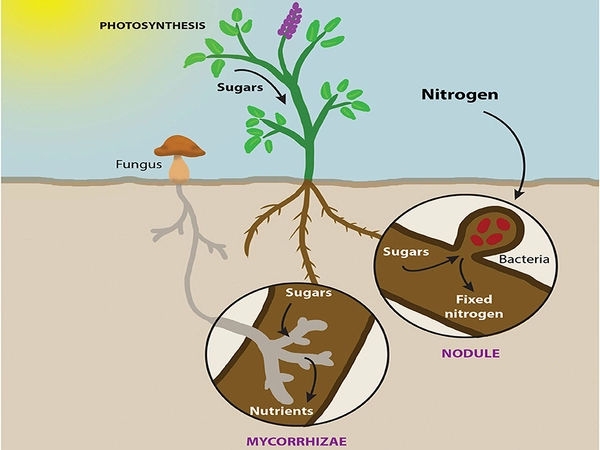
The researchers discovered a core microbiome in Miscanthus that comprised nitrogen-fixing bacteria previously discovered in rice and barley. All of these bacteria contribute to the acquisition of nitrogen, a necessary nutrient for plant growth. Recruiting nitrogen-fixing microorganisms may help plants adapt to varied conditions, but more crucially, this capability helps to the grass’s long-term viability as a bioenergy crop.
On the other hand, the effects of genetic variation across plants had a smaller effect on the rhizosphere microbiome, which was more significantly influenced by the soil. Nonetheless, the plants placed a larger priority on attracting fungal than other microorganisms.
The researchers are interested in parsing out which genes play a role in influencing the microbiome. “The microsatellites do not have a biological function and are not representative of the whole genome. It would be nice if we could sequence the whole Miscanthus genome and figure out how the genes affect nitrogen fixation,” Ji said.
“Crop breeding is based on yield. However, we must adopt a broader perspective and assess how bacteria can assist to crop sustainability,” Kent stated. “The attractiveness of dealing with wild plants is that there is a great deal of genetic variety to study. We can determine which varieties are effective in recruiting nitrogen-fixing bacteria because these crops require fewer fertilizers. It’s an intriguing possibility as we begin to alter these plants for bioenergy use.”