Synthetic biology brings together engineers and biologists to design and build novel biomolecular components, networks, and pathways, and then use these constructs to rewire and reprogram organisms. These re-engineered organisms will change our lives in the coming years, resulting in cheaper drugs, “green” fuel for our cars, and targeted therapies for combating “superbugs” and diseases like cancer.
Synthetic biology aims to achieve robust control of biological processes in order to create designer organisms for a variety of industrial, diagnostic, and therapeutic applications. Researchers at Aarhus University’s iNANO center have created RNA origami sponges and CRISPR-based regulators for advanced genetic control of enzymatic pathways in microorganisms in order to improve the production of valuable biochemicals.
One of the main pillars of the now mature field of synthetic biology has been the development of tools for precise control of biological processes. These scientific tools take principles from a variety of research fields and combine them to create unique applications that have the potential to transform modern society.
Because of their compatibility with folding and expression in cells, modern RNA nanotechnology innovations have enormous potential in the biological context, but they also pose unique challenges, such as tight performance conditions and the inherent instability of RNA molecules.
The transcription factors activated gene expression by targeting RNA scaffolds to promoter regions. It was discovered that the orientation of the scaffold and the number of transcription factors recruited can influence the expression strength.
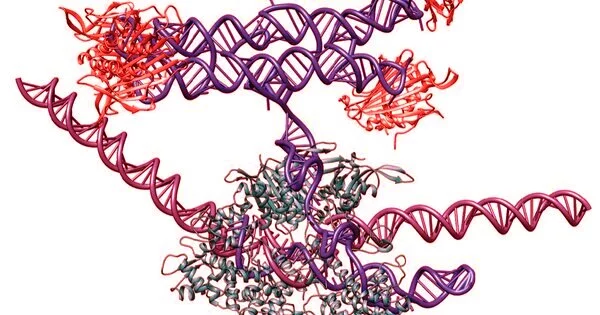
However, a recent structural RNA design approach developed in the Andersen lab, termed ‘RNA origami’, is trying to tackle this. This approach attempts to generate complex human-made RNA-based devices that are stable in cells, interact with other biomolecules, including other RNA and proteins, and enable unique applications, particularly in the context of gene regulation.
Demonstrated by two distinct approaches recently published, RNA origami is presented as a sophisticated RNA design platform that when applied in the cellular context, generates unique molecules for synthetic biology-based regulation.
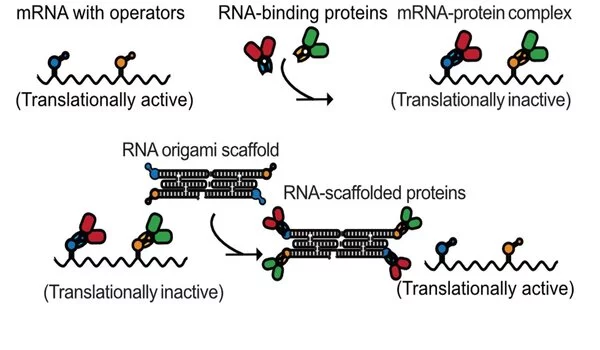
RNA sponges regulate enzyme production in bacteria
In the first approach, RNA origami was used to achieve precise control of protein production levels when expressed in bacteria. Self-inhibiting protein expression cassettes were made by installing a strong binding site for the expressed protein in its own gene. Afterwards, RNA origami decorated with the same protein-binding sites was expressed in large excess.
In this way, the RNA origami serves as a protein sponge that sequesters proteins in the cell and allows expression of the self-inhibited protein. This general concept was shown to enable regulation of several proteins simultaneously and turn on enzymatic pathways for improved product yields.
CRISPR-based regulators for yeast chemical factories
In the second approach, RNA origami was combined with CRISPR, one of the most popular modern molecular biology techniques, to regulate gene expression in yeast. The RNA origamis were incorporated into small RNAs that direct CRISPR-Cas9 to target specific sequences in the DNA genome. Protein-binding sites capable of attracting transcription factors were added to the RNA origami scaffolds.
The transcription factors activated gene expression by targeting RNA scaffolds to promoter regions. It was discovered that the orientation of the scaffold and the number of transcription factors recruited can influence the expression strength. Finally, it was demonstrated that multi-enzyme pathways could be controlled for high-yield production of the anti-cancer drug violacein.