Individual mRNAs in bacterial cells can live for seconds to more than an hour. However, the lifetime of bacterial mRNA ranges between 1 and 3 minutes, making it far less stable than eukaryotic mRNA. mRNA lifespan in mammalian cells varies from a few minutes to several days. Eukaryotic mRNAs have a lifespan of several hours, but E. coli mRNAs have a lifespan of no more than five seconds. Pre-mRNAs are initially coated in RNA-stabilizing proteins, which keep the pre-mRNA from degrading while it is processed and released from the nucleus.
New research sheds light on how mRNAs are formed and how they influence protein creation within our cells once they reach maturity. The discoveries have ramifications not only for developing effective vaccine doses, but also for determining the biological foundations of numerous malignancies and disorders.
It took a global pandemic, but the importance of messenger RNA in many aspects of life has been highlighted in the last year with the successful rollout of mRNA vaccines to battle the SARS-Cov-2 virus.
Wendy Gilbert’s team at Yale offers light on how mRNAs are formed and how they influence protein creation inside our cells after they reach maturity in two recent publications released. The discoveries have ramifications not only for developing effective vaccine doses, but also for determining the biological foundations of numerous malignancies and disorders.
Cells precisely copy or transcribe genes encoded in DNA into mRNAs, which then convey those instructions to the ribosome, the machinery within the cell that produces the proteins that carry out practically all life tasks.
Wendy Gilbert
“It’s been wonderful to explore the beginning and conclusion of this process,” said Gilbert, an associate professor of molecular biophysics and biochemistry. Cells precisely copy or transcribe genes encoded in DNA into mRNAs, which then convey those instructions to the ribosome, the machinery within the cell that produces the proteins that carry out practically all life tasks, according to textbook biology.
Because of this critical role, mRNA has been a prominent research target for decades, including research that has resulted in the rapid creation of mRNA vaccines in the fight against COVID-19. Pfizer and Moderna’s vaccines contain mRNA-based instructions for cells to create proteins that detect spike proteins on the surface of the SARS-Cov-2 virus, making them targets for immune system destruction.
While RNA is made up of only four bases, or nucleotides, intricate biochemical interactions with other molecules can affect its structure and function. Pseudouridine, an isomer whose presence is critical to the success of mRNA vaccines, is one such chemical that alters mRNA. Gilbert’s lab detected the presence of pseudouridine in normal cellular mRNA long before the pandemic. She was curious at the time about how these mRNA alterations are formed and how they affect the mRNA’s function.
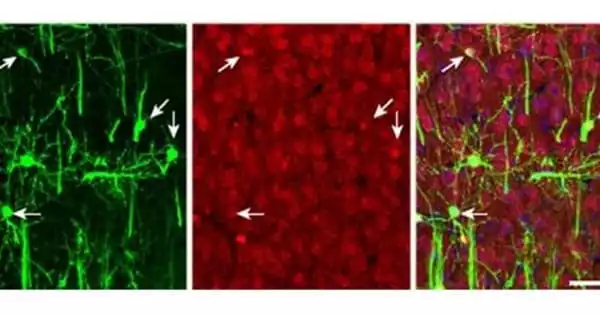
A team led by Nicole Martinez, a postdoctoral fellow in Gilbert’s group, discovered that pseudouridine plays an important role in the creation of mRNAs in one of the new investigations. The researchers discovered pseudouridine at the earliest stages of mRNA synthesis. And the researchers identified evidence that it directs the splicing of genetic material into mRNAs, which in turn govern gene activity, as they publish in the journal Molecular Cell.
These findings give fresh light on the causes of diseases connected to pseudouridine variations, such as mitochondrial myopathy, digestive issues, intellectual impairment, and viral infection resistance. Several tumors, for example, have been related to higher levels of pseudouridine, implying that incorrect mRNA splicing may cause tumor growth and cancer metastasis.
Yale researchers studied how ribosomes determine how many proteins to generate based on the genetic instructions they receive from mRNAs in a second publication published in the journal Cell Systems. A team lead by Rachel Niederer, an associate research scientist in Gilbert’s lab, created a new approach called direct analysis of ribosome targeting (DART) to uncover regulatory elements that can stimulate and inhibit ribosomal protein production. Manipulation of such components in mRNAs – in this case, within yeast – enabled scientists to alter protein output by a thousand-fold, they report.
According to the researchers, the ability to precisely modify protein production has direct applications in altering doses of mRNA vaccines such as those used to combat COVID. Pfizer awarded the Gilbert lab and Carson Thoreen, associate professor of cellular and molecular physiology at Yale, a $1.7 million grant for their studies. Gilbert, on the other hand, emphasized that the method might be applied to the development of any protein-based therapeutics for a wide range of ailments.