Researchers at Children’s Hospital of Philadelphia (CHOP), St. Jude Children’s Research Hospital (St. Jude), and the Children’s Oncology Group (COG) today announced a significant paradigm shift in the understanding of T-lineage acute lymphoblastic leukemia (T-ALL), an aggressive and high-risk form of cancer, to one that is frequently caused by genetic changes in non-coding portions of our DNA. The collaborative study, funded by the Gabriella Miller Kids First Pediatric Research Program (Kids First) and the National Institutes of Health (NIH) Common Fund, was published today in the journal Nature.
T-ALL patients in children, teens, and young adults typically react favorably to first treatment. Patients who relapse or have disease that is resistant to treatment, on the other hand, frequently have a poor prognosis. Given the disease’s aggressive character and rapid progression, as well as a lack of understanding of its genetic foundation, experts recognized the urgent need for novel and effective techniques to diagnosis and therapy.
“This paper is the first to transcend previous barriers and comprehensively profile the whole genome, uncovering critical insights in more than 1,300 children, adolescents and young adults with T-ALL,” said David T. Teachey, MD, an attending physician, Director of Clinical Research at the Center for Childhood Cancer Research at CHOP and Chair of the Acute Lymphoblastic Leukemia disease committee in the COG. “These findings are a significant clinical advancement, as the goal in treating T-ALL is to prevent relapse, which requires identifying the patients most at risk. This data now makes it possible to risk stratify patients with T-cell leukemia, identifying those with a high-risk of relapsing so we can treat them with newer or alternative medicines.”
This paper is the first to transcend previous barriers and comprehensively profile the whole genome, uncovering critical insights in more than 1,300 children, adolescents and young adults with T-ALL.
David T. Teachey
Previous studies were unable to discover significant genetic changes in T-ALL because they focused on the coding genome, the section of DNA that encodes proteins, which are the building blocks of cells. However, only 1% of DNA is coding, with the remaining 99% being non-coding.
Scientists now understand that the non-coding area has an important role in regulating biological processes, despite previously believing it to be meaningless. It tells the cell when to make specific proteins, much like a crossing guard directing people safely across the street.
In this case, researchers studied more than 1,300 patients treated on the COG AALL0434 clinical trial and sequenced both the tumor and non-tumor genomes of each patient. While the researchers previously suspected that non-coding DNA in T-ALL played an important role, this study’s findings are the first ever to establish that at a large scale.
The study found that approximately 60% of the genetic changes driving T-ALL cancer cells are non-coding changes. This fundamentally alters the way researchers think about T-ALL, offering a better understanding of disease biology. This leads to innovative treatments, including new immunotherapies developed at CHOP and St. Jude.
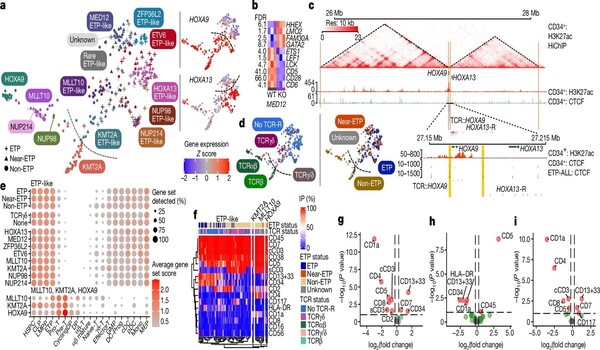
Traditionally, individuals with T-All have been classified by risk based on their therapeutic response and immunophenotype, which includes profiling cell surface proteins as part of the diagnostic workup. While cell surface protein expression helps characterize T-ALL subtypes, it has not consistently identified patients with a good prognosis. The new complete data indicated why, strongly indicating that a genomic method should replace the current immunophenotypic classification system. As a result, the researchers created models that use genetics and therapy response to appropriately risk stratify patients with T-ALL, and they are currently testing the results using patient samples from the forthcoming T-ALL COG study.
“It was striking how abundant these non-coding changes were and how many of them were enhancer perturbation events, whether it was hijacking or co-option of an existing enhancer, or changes that generated a new enhancer,” said Charles Mullighan, MBBS, MD, St. Jude Children’s Research Hospital, Comprehensive Cancer Center deputy director and Department of Pathology member. “We now have a much stronger framework to take these alterations back to the lab and say now we’ve got better information to build the right models to understand the biology, and then to test therapy. We have very clear information that these are the sorts of alterations that people need to focus on to build a diagnostic test.”
T-ALL was classified into 15 subtypes based on unique gene expression and genetic causes, including previously unidentified subtypes. They revised the classification of recognized subtypes and demonstrated that driver lesions, other genetic alterations, and the original cell type all contribute to defining a condition’s genomic subtype as well as its clinical and biological characteristics. They also discovered a strong relationship between the type of gene changes and T-ALL outcomes. This new discovery demonstrates that not only whatever gene is mutated in cancer cells, but also how it is altered, helps define prognosis and chances of cure.
“Future research must continue to determine broader applications for this approach,” said Teachey. “These findings offer a strong a roadmap for improving patient outcomes and curing more children and adults with T-ALL.”