Mammals evolved from members of the reptilian order Therapsida during the Triassic Period (about 252 million to 201 million years ago). Therapsids, which included Lystrosaurus, were mammal-like reptiles that thrived during the early Triassic Period (252 million to 201 million years ago).
A new study published in Nature provides the most detailed timeline of mammal evolution to date. The study describes a new and fast computational method for obtaining precisely dated evolutionary trees, known as ‘timetrees.’ The authors applied the novel method to a mammal genomic dataset to answer a long-standing question about whether modern placental mammal groups evolved before or after the Cretaceous-Palaeogene (K-Pg) mass extinction, which wiped out over 70% of all species, including all dinosaurs.
The findings confirm that the ancestors of modern placental mammal groups predate the K-Pg extinction 66 million years ago, putting an end to a debate about the origins of modern mammals. Placental mammals are the most diverse group of living mammals, including primates, rodents, cetaceans, carnivorans, chiropterans (bats), and humans.
Dr. Mario dos Reis (Queen Mary University of London) and Professor Phil Donoghue (University of Bristol) led the research team, which included scientists from Queen Mary, the University of Bristol, UCL, Imperial College London, and the University of Cambridge.
Our data pipeline gathered as much genomic data for as many mammal species as possible. This was difficult because genetic databases contain inaccuracies, and we needed to devise a strategy for identifying low-quality samples or mislabeled data that needed to be removed.
Dr. Asif Tamuri
Dr. Sandra lvarez-Carretero, the paper’s lead author from UCL (then at Queen Mary), says: “We were able to reduce uncertainties and obtain a precise evolutionary timeline by incorporating complete genomes in the analysis as well as the necessary fossil information. Did modern mammal groups coexist with dinosaurs, or did they evolve after the dinosaurs died out? We now have a conclusive answer.”
“One of the most contentious topics in evolutionary biology is the timeline of mammal evolution. Early studies estimated the origin of modern placental groups deep in the Cretaceous, during the dinosaur era. Over the last two decades, studies have alternated between post- and pre-K-Pg diversification scenarios. Our precise timeline solves the problem.” Prof. Donoghue, the paper’s co-senior author, adds.
With global sequencing projects now producing hundreds to thousands of genome sequences and plans to sequence more than a million species in the near future, evolutionary biologists will soon have a wealth of data at their disposal. Current methods for analyzing the vast genomic datasets available and creating evolutionary timelines, on the other hand, are inefficient and computationally expensive.
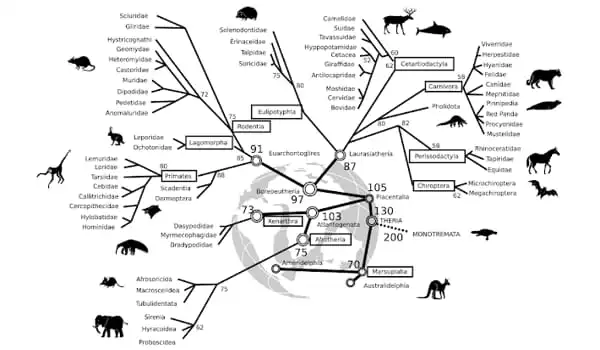
“A fundamental goal of biology is to infer evolutionary timelines. However, cutting-edge methods rely on computers to simulate evolutionary timelines and evaluate the most plausible ones. This was difficult in our case due to the massive dataset examined, which included genetic data from nearly 5,000 mammal species and 72 complete genomes” According to Dr. dos Reis.
In this study, the researchers developed a new, fast Bayesian approach for analyzing large numbers of genome sequences while accounting for data uncertainties. “We overcame the computational challenges by breaking the analysis down into sub-steps: first simulating timelines using the 72 genomes, then using the results to guide simulations on the remaining species. Using genomes reduces uncertainty by allowing simulations to reject implausible timelines” Dr. dos Reis says.
“Our data pipeline gathered as much genomic data for as many mammal species as possible. This was difficult because genetic databases contain inaccuracies, and we needed to devise a strategy for identifying low-quality samples or mislabeled data that needed to be removed “Dr. Asif Tamuri, co-lead author of the paper from UCL, who was in charge of compiling the mammal genomic dataset, adds.
Using their novel approach, the team was able to reduce the computation time for this complex analysis from decades to months. “We would have had to wait decades to infer the mammal timetree if we had tried to analyze this large mammal dataset on a supercomputer without using the Bayesian method we developed. Consider how long this analysis could take if we used our own PCs “explains Dr. lvarez-Carretero “In addition, we were able to cut computation time by a factor of 100. This new approach not only allows for the analysis of genomic datasets, but it also significantly reduces CO2 emissions due to computing by being more efficient” Dr. lvarez-Carretero continues.
The method developed in the study could be applied to other contentious evolutionary timelines that require large dataset analysis. The idea of estimating a reliable evolutionary timescale for the Tree of Life now appears within reach by combining the novel Bayesian approach with the upcoming genomes from the Darwin Tree of Life and Earth BioGenome projects.