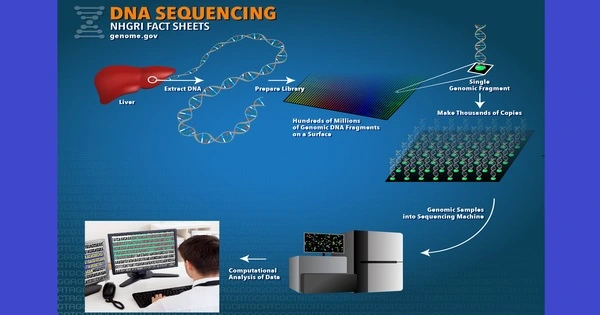
DNA sequencing is the technique of determining the nucleic acid sequence – the order of nucleotides in DNA. It is a method for determining the precise order of nucleotides (adenine, cytosine, guanine, and thymine) in a DNA molecule. It comprises any method or technology used to establish the order of the four bases: adenine, guanine, cytosine, and thymine. The development of fast DNA sequencing tools has considerably advanced biological and medical research and discoveries. This information is crucial for interpreting genetic codes, identifying genes, analyzing genetic variants, and a variety of applications in domains like as genetics, genomics, medicine, and forensics.
Basic biological research, DNA Genographic Projects, and several applied sectors such as medical diagnosis, biotechnology, forensic biology, virology, and biological systematics have all made knowledge of DNA sequences vital. Comparing healthy and altered DNA sequences can be used to detect many diseases, including cancer, describe antibody repertoires, and guide patient treatment. Having a speedy way to sequence DNA allows for more tailored medical care to be delivered, as well as the identification and cataloging of more organisms.
There are several methods of DNA sequencing, with the most commonly used being:
- Sanger Sequencing (Chain Termination Method): This was the first commonly used DNA sequencing method. DNA polymerase, DNA primers, and chain-terminating nucleotides (dideoxynucleotides) are used. When the polymerase adds nucleotides to a developing DNA strand, it may include a chain-terminating nucleotide rather than a conventional one. This produces a succession of variable length fragments that are separated by electrophoresis and read to determine the DNA sequence.
- Next-Generation Sequencing (NGS): NGS technology have transformed DNA sequencing by allowing for high-throughput, parallel sequencing of millions of DNA fragments at the same time. Illumina, Ion Torrent, and PacBio are three common NGS platforms. NGS uses a variety of approaches, including Illumina sequencing (based on synthesis), Ion Torrent sequencing (based on pH changes during nucleotide incorporation), and PacBio sequencing (based on single molecule, real-time sequencing).
- Third-Generation Sequencing: These methods aim to address some limitations of NGS, such as short read lengths. One notable example is Single-Molecule Real-Time Sequencing (SMRT) by Pacific Biosciences (PacBio), which reads longer DNA fragments in real-time. Another is nanopore sequencing, developed by Oxford Nanopore Technologies, which involves threading DNA through a nanopore and measuring changes in electrical current as each nucleotide passes through.
- Metagenomic Sequencing: This technique is used to study the DNA of entire microbial communities present in environmental samples or in the human body. It can help identify various species and understand their genetic diversity.
- Targeted Sequencing: Rather than sequencing the entire genome, this method sequences only specific regions of interest in the genome. It is frequently used for activities such as finding alterations in cancer genes and researching certain genetic markers.
DNA sequencing has had a significant impact on a wide range of scientific and medical sectors, including genetics, personalized medicine, evolutionary biology, and disease research. It has become more affordable and accessible over time, making it a valuable tool for academics and physicians.