AI technology can certainly play a role in generating novel proteins from scratch. This process is often referred to as “de novo protein design”. By using machine learning algorithms and high-performance computing, AI can be used to predict the 3D structure and stability of novel proteins, which can then be synthesized in the laboratory for further testing and characterization.
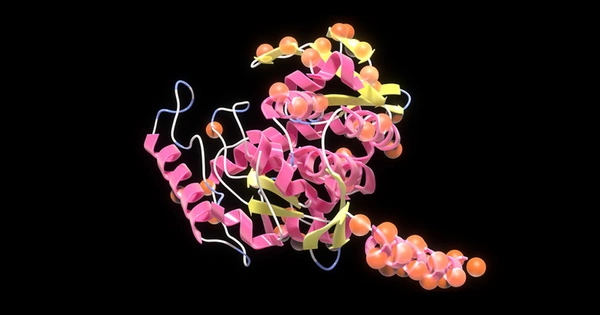
Scientists have developed an artificial intelligence system capable of creating artificial enzymes from scratch. Some of these enzymes performed as well as those found in nature in laboratory tests, even when their artificially generated amino acid sequences diverged significantly from any known natural protein.
The experiment shows that natural language processing, despite being designed to read and write language text, can learn at least some biological principles. ProGen is an AI program developed by Salesforce Research that uses next-token prediction to assemble amino acid sequences into artificial proteins.
The new technology, according to scientists, has the potential to be more powerful than directed evolution, the Nobel Prize-winning protein design technology, and it will energize the 50-year-old field of protein engineering by speeding the development of new proteins that can be used for almost anything, from therapeutics to plastic degradation.
When you train sequence-based models with a large amount of data, they are extremely powerful in learning structure and rules. They learn what words can co-occur and how to compose sentences.
Nikhil Naik
“The artificial designs perform much better than designs inspired by the evolutionary process,” said James Fraser, Ph.D., professor of bioengineering and therapeutic sciences at the UCSF School of Pharmacy and one of the study’s authors.
“The language model is learning aspects of evolution, but it’s different than the normal evolutionary process,” Fraser said. “We now have the ability to tune the generation of these properties for specific effects. For example, an enzyme that’s incredibly thermostable or likes acidic environments or won’t interact with other proteins.”
Scientists simply fed the amino acid sequences of 280 million different proteins of various types into the machine learning model and let it digest the information for a couple of weeks to create the model. The model was then fine-tuned by priming it with 56,000 sequences from five lysozyme families, as well as contextual information about these proteins.
The model quickly generated a million sequences, and the research team chose 100 to test based on how closely they resembled natural protein sequences, as well as how naturalistic the underlying amino acid “grammar” and “semantics” of the AI proteins were.
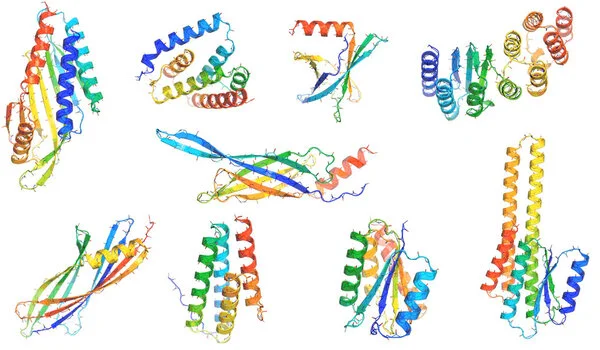
Tierra Biosciences screened 100 proteins in vitro and created five artificial proteins to test in cells, comparing their activity to hen egg white lysozyme, an enzyme found in the whites of chicken eggs (HEWL). Similar lysozymes are found in human tears, saliva, and milk, where they protect against bacteria and fungi.
Two of the artificial enzymes were able to break down bacterial cell walls with activity comparable to HEWL, despite the fact that their sequences were only about 18% identical. The two sequences were approximately 90% and 70% identical to any known protein, respectively.
Just one mutation in a natural protein can make it stop working, but in a different round of screening, the team found that the AI-generated enzymes showed activity even when as little as 31.4% of their sequence resembled any known natural protein.
Simply by studying the raw sequence data, the AI was able to learn how the enzymes should be shaped. The atomic structures of the artificial proteins were measured using X-ray crystallography and looked exactly as they should, despite the fact that the sequences were unlike anything seen before.
Salesforce Research created ProGen in 2020, based on a type of natural language programming developed by their researchers to generate English language text. They knew from previous work that the AI system could teach itself grammar and word meaning, as well as other underlying rules that contribute to well-written writing.
“When you train sequence-based models with a large amount of data, they are extremely powerful in learning structure and rules,” said Nikhil Naik, Ph.D., Director of AI Research at Salesforce Research and the paper’s senior author. “They learn what words can co-occur and how to compose sentences.”
The possibilities for protein design were nearly limitless. Lysozymes are small proteins, containing up to 300 amino acids. However, with 20 possible amino acids, there are an enormous number of possible combinations (20300). That is more than all the humans who have lived throughout history multiplied by the number of grains of sand on Earth multiplied by the number of atoms in the universe.