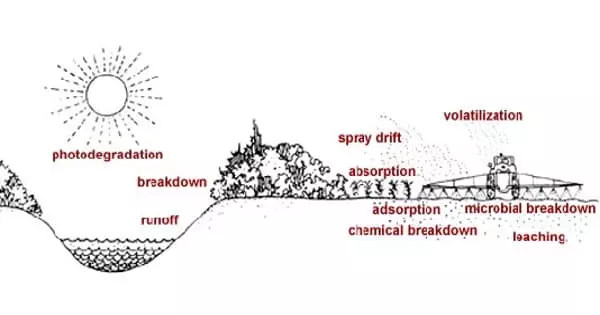
Researchers first learned that plants can detect carbon dioxide (CO2) concentrations more than 50 years ago. Stomata, or “breathing” pores in leaves, open and close in response to changes in CO2 levels, regulating water evaporation, photosynthesis, and plant growth. More than 90% of the water in plants evaporates through the stomata.
Due to the increased impacts of carbon dioxide on the climate and water supplies in a warming world, the regulation of stomatal pore openings by CO2 is essential for deciding how much water plants lose.
The location of the carbon dioxide sensor and an explanation of how it functions in plants, however, have long been a mystery.
Scientists at the University of California, San Diego recently made a significant advancement in the identification of the long-sought CO2 sensor in Arabidopsis plants and unraveled its functional components.
UC San Diego project scientist Yohei Takahashi, School of Biological Sciences Distinguished Professor Julian Schroeder and their colleagues identified the CO2 sensor mechanism and detailed its genetic, biochemical, physiological and predicted structural properties. Their results are published December 7 in Science Advances.
Stomatal pores regulate plant water loss, hence the sensor is essential for water management and has implications for managing drought brought on by climate change, wildfires, and agricultural crops.
“For each carbon dioxide molecule taken in, a typical plant loses some 200 to 500 water molecules to evaporation through the stomatal pores,” said Schroeder, Novartis Chair and faculty member in the Department of Cell and Developmental Biology. “The sensor is extremely relevant because it recognizes when CO2 concentrations go up and determines how much water a plant loses as carbon dioxide is taken in.”
Our findings reveal that plants sense changes in CO2 concentration by the reversible interaction of two proteins to regulate stomatal movements. This could provide us a new plant engineering and chemical target towards efficient plant water use and CO2 uptake from the atmosphere.
Yohei Takahashi
The composition of the sensor was a key discovery from the recent research. The researchers discovered that the sensor functions as a result of two plant proteins cooperating, rather than linking it to a single source or protein. These were identified as 1) a “high leaf temperature1” protein kinase known as HT1 and 2) specific members of a mitogen-activated protein kinase family, or “MAP” kinase enzyme, known as MPK4 and MPK12.
“Our findings reveal that plants sense changes in CO2 concentration by the reversible interaction of two proteins to regulate stomatal movements,” said Takahashi, who is now based at the Institute of Transformative Bio-Molecules, Japan. “This could provide us a new plant engineering and chemical target towards efficient plant water use and CO2 uptake from the atmosphere.”
The team’s discoveries, which have been documented in a UC San Diego patent, may inspire improvements in how well plants use water as CO2 levels rise.
“This finding is relevant for crops but also for trees and their deep roots that can dry out soils if there’s no rain for long periods, which can lead to wildfires,” said Schroeder. “If we can use this new information to help trees respond better to increases in CO2 in the atmosphere, it’s possible they would more slowly dry out the soil. Similarly, the water use efficiency of crops could be improved more crop per drop.”
The researchers worked with professor Andrew McCammon from the Department of Chemistry and Biochemistry as well as graduate student Christian Seitz to further investigate their sensor discoveries. Seitz and McCammon developed a thorough model of the complex sensor structure using cutting-edge methods.
The model identified regions in which genetic alterations are known to limit a plant’s capacity to control transpiration in response to carbon dioxide. The updated imaging revealed that the mutants congregate along the interface between the two sensor proteins, HT1 and MPK.
“This work is a wonderful example of curiosity-driven research that brings together several disciplines from genetics to modeling to systems biology and results in new knowledge with the ability to aid society, in this case by making more robust crops,” said Matthew Buechner, a program director in the U.S. National Science Foundation’s Directorate for Biological Sciences, which supported the research.
The paper’s full author list: Yohei Takahashi, Krystal Bosmans, Po-Kai Hsu, Karnelia Paul, Christian Seitz, Chung-Yueh Yeh, Yuh-Shuh Wang, Dmitry Yarmolinsky, Maija Sierla, Triin Vahisalu, J. Andrew McCammon, Jaakko Kangasjarvi, Li Zhang, Hannes Kollist, Thien Trac and Julian I. Schroeder.