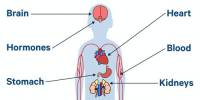
According to a recent study, close family consanguinity unions may raise the risk of common illnesses including type 2 diabetes and post-traumatic stress disorder (PTSD).
Using a novel technique that lessens confounding caused by sociocultural factors, researchers from the Wellcome Sanger Institute and their collaborators at Queen Mary University of London analyzed the genomic data of various groups to investigate the association between autozygosity, a measure of genetic relatedness between a person’s parents, and the prevalence of common diseases.
They focused their analysis on the Genes & Health cohort, which consists of British individuals of Pakistani and Bangladeshi descent, as well as individuals of both European and South Asian descent from the UK Biobank.
In collaboration with the researchers, the Genes & Health Community Advisory Board created a paper that can be accessed by the general public and explains the goals, procedures, and outcomes of the study.
The findings, published in Cell, help shed light on the complex interplay between genetics and health outcomes, especially among populations with higher rates of consanguinity.
The social and cultural practice of consanguinity is the union of two blood relatives who have a recent common ancestor in common, such as a grandparent or great-grandparent. This practice is observed across the world with varying prevalence. Over 10% of the global population consists of individuals who are the offspring of second cousins or closer. In the UK, consanguinity is more common among some British South Asian communities.
Consanguinity increases autozygosity, or the percentage of a person’s DNA that is inherited identically from both parents. Consanguinity increases the risk of rare single-gene disorders by raising the likelihood that a person may inherit the same uncommon DNA alteration in a disease-causing “recessive” gene, although its effects on common diseases are still not fully understood.
British Pakistanis and Bangladeshis have higher rates of several diseases than the UK average for example a four-to-six-fold increased risk of developing type 2 diabetes compared to individuals of European ancestry.
The findings have the potential to inform disease risk prediction as well as future research efforts to identify specific genetic variants associated with these diseases, not only within these specific communities but also globally, particularly across populations where consanguinity rates are higher. This could be used to help stratify individuals for earlier screening and identify potential drug targets.
Dr. Hilary Martin
However, these diseases involve a complex interplay of genetic and environmental factors, and, prior to this study, it was unknown whether consanguinity plays a role.
In this new study, researchers from the Wellcome Sanger Institute and their collaborators set out to assess the impact of consanguinity on complex genetic diseases.
The teams analyzed genomic data to describe different patterns of consanguinity in distinct populations, including 23,978 British individuals of Pakistani and Bangladeshi descent from the Genes & Health cohort, and 397,184 individuals of European or South Asian descent from the UK Biobank cohort.
They found that ~33% of individuals in Genes & Health were offspring of second cousins or closer, versus 2% of individuals of European descent in UK Biobank.
They then investigated the relationship between autozygosity and the prevalence of common diseases. For this, they restricted their analysis to a set of ~5,700 individuals in Genes & Health and UK Biobank with parents who were inferred to be first cousins based on the genetic data.
The precise level of autozygosity within this defined “highly consanguineous” group is determined randomly, ranging from 4 to 15%, and the researchers demonstrated that it is unrelated to sociocultural and environmental factors, such as religion, education, or diet, which may also have an impact on health traits. This innovative approach made sure that any biological causes, rather than confounding factors, were behind any observed connections between autozygosity and illnesses.
Among the 61 complex genetic diseases examined in the Genes & Health and UK Biobank cohorts, researchers identified 12 diseases and disorders associated with increased autozygosity resulting from consanguinity. These included type 2 diabetes, asthma, and PTSD. The associations with type 2 diabetes and PTSD were then validated in a separate dataset from the consumer genetic company 23andMe Inc., using a between-sibling analysis technique.
Analysis suggested that consanguinity may account for approximately 10% of type 2 diabetes cases among British Pakistanis and around 3% of cases among British Bangladeshis.
Consanguinity may have certain health hazards, but they should be weighed against the practice’s good social effects and other, more significant modifiable risk factors like exercise, smoking, and body mass index.
This research reveals important insights into the factors influencing health outcomes and the associations between autozygosity and complex diseases within British Pakistani and Bangladeshi communities. It recommends that in order to identify particular variants and genes with recessive effects, genetic studies of complex disorders should be expanded.
Daniel Malawsky, first author of the study and Ph.D. student at the Wellcome Sanger Institute, said, “While consanguinity has a smaller role in common diseases compared to other factors, it is still essential to understand its specific influence on health in these communities. Our new method exploring the natural variation in expected autozygosity among offspring of first cousins was a key breakthrough in helping us to test its impact.”
“Some of our results suggested that cultural and environmental factors associated with consanguinity can sometimes exaggerate associations between autozygosity and health-related traits, or even mask truly causal associations. Our results suggest that some findings from previous studies linking autozygosity to complex traits in humans may have been misleading.”
Cllr Ahsan Khan, chair of the Genes & Health community advisory board and councilor at Waltham Forest, said, “This work underscores the significance of culturally sensitive approaches in health research, acknowledging the delicate balance between social benefits and any potential risks.”
“The research team actively engaged community members, taking into account our traditions, cultures, and religious practices. By empowering people with the knowledge to make informed health decisions, we can help tackle the health disparities in our communities, especially in diseases like type 2 diabetes.”
Prof Sarah Finer, author of the study, co-lead of the Genes & Health research program from Queen Mary, University of London, said, “This research would not have been possible without the many thousands of volunteers who generously agreed to participate in the Genes & Health study and UK Biobank.”
Dr. Hilary Martin, senior author of the paper and group leader at the Wellcome Sanger Institute, said, “The findings have the potential to inform disease risk prediction as well as future research efforts to identify specific genetic variants associated with these diseases, not only within these specific communities but also globally, particularly across populations where consanguinity rates are higher. This could be used to help stratify individuals for earlier screening and identify potential drug targets.”
Researchers conducted a within-sibling analysis using the consumer genetics company 23andMe Inc. cohort. This involved 545,806 research-consented individuals with at least one genetically-inferred full sibling also in the cohort, using self-reported phenotypes. This method of between-sibling analysis is regarded as the industry standard for determining the causal relationship between genetic variables.