Antibiotic-resistant microorganisms cause illness in about 670,000 individuals in Europe each year, and 33,000 of them pass away from the diseases. Pathogens that are resistant to several, or even all, existing antibiotics are particularly dreaded. One of these is the bacterium Acinetobacter baumannii, feared today above all as the “hospital superbug.”
According to estimates, this pathogen alone is responsible for up to 5% of all hospital-acquired infections and 10% of all bacterial infections that result in mortality. This puts A. baumannii right at the top of a list of pathogens for which according to the World Health Organization (WHO) there is an urgent need to develop new therapies.
Understanding which characteristics make A. baumannii a pathogen is one of the prerequisites for this. To this end, bioinformaticians led by Professor Ingo Ebersberger of Goethe University Frankfurt and the LOEWE Center for Translational Biodiversity Genomics (LOEWE-TBG) are comparing the genomes and the proteins encoded therein across a wide range of different Acinetobacter strains.
The distinctions between harmful and harmless strains can be used to make inferences about which genes contribute to pathogenicity.
Studies in this area have thus far focused on determining whether a gene is present in a particular bacterial strain due to a lack of adequate methodologies. This, however, ignores the reality that genes, and thus the proteins they encode, can be altered in order for bacteria to acquire new traits.
In this way, drastic functional modifications can be achieved over short evolutionary time spans. We assume that bacterial strains that differ in terms of their T4A pili also interact differently with their environment. This might determine, for example, in which corner of the human body the pathogen settles.
Professor Ingo Ebersberger
Due to this, Ebersberger’s team created a bioinformatics method to track the modification of proteins along an evolutionary lineage. In collaboration with microbiologists from the Institute for Molecular Biosciences and the Institute of Medical Microbiology and Infection Control at Goethe University Frankfurt, they have now applied this method to Acinetobacter for the first time. The research is published in the journal PLOS Genetics.
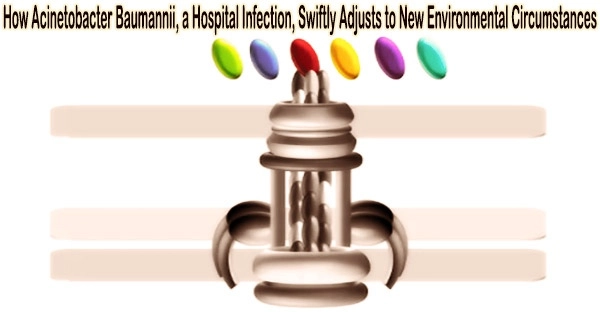
In the process, the researchers concentrated on hair-like cell appendages, known as type IVa (T4A) pili, which are prevalent in bacteria and that they use to interact with their environment.
The fact that the T4A pili are found in harmless bacteria and have even been discovered as a crucial component of the virulence of some diseases implies that they have continually picked up new traits related to pathogenicity during evolution.
The study team was able to demonstrate that the group of pathogenic Acinetobacter strains exhibits obvious alterations in the protein ComC, which is located on the tip of the T4A pili and is crucial for their function.
Even different strains of A. baumannii have different variants of this protein. This prompts bioinformatician Ebersberger to compare the T4A pili to a multipurpose gardening tool with a constant handle and replaceable attachments.
“In this way, drastic functional modifications can be achieved over short evolutionary time spans,” Ebersberger said. “We assume that bacterial strains that differ in terms of their T4A pili also interact differently with their environment. This might determine, for example, in which corner of the human body the pathogen settles.”
The aim is to use this knowledge of the unexpectedly high diversity within the pathogen to improve the treatment of A. baumannii infections, as Ebersberger explains, “Building on our results, it might be possible to develop personalized therapies that are tailored to a specific strain of the pathogen.”
The study by Ebersberger and his associates also demonstrates another fact: Prior investigations into the comparative genomes of A. baumannii are likely to have barely touched the surface.
“Our approach has gone a long way towards resolving the search for possible components that characterize pathogens,” says Ebersberger.





![Report on Beximco Knitting Ltd [ Part-1]](https://assignmentpoint.com/wp-content/uploads/2013/04/Beximco-Knitting-Limited-110x55.jpg)