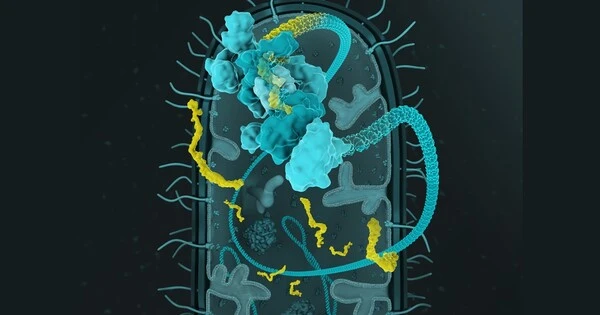
Researchers have given gut bacteria data logger functionality in order to track which genes are active in the bacteria. These microorganisms may one day provide a noninvasive method of diagnosing disease or evaluating the impact of diet on health.
Numerous bacteria live in our gut and aid in digestion. But, exactly, what do microorganisms do inside the body? When and which enzymes do they produce? And how do bacteria metabolize health-promoting foods that keep us healthy?
To find answers to such questions, researchers at ETH Zurich’s Department of Biosystems Science and Engineering in Basel modified bacteria to act as data loggers for gene activity. They have now tested these bacteria in mice in collaboration with scientists from the University Hospital of Bern and the University of Bern. This is an important step toward the future use of sensor bacteria in medicine for applications such as diagnosing malnutrition and determining which diets are best for an individual.
Immune system becomes data logger
Researchers led by Randall Platt, Professor of Biological Engineering at ETH Zurich, worked on the data logger function for several years. They used the CRISPR-Cas mechanism, which is a type of immune system found in many bacterial species, to accomplish this. If the bacteria are attacked by viruses, they can incorporate viral DNA or RNA snippets into a section of their own genome known as the CRISPR array. This allows the bacteria to “remember” viruses with which they have come into contact, allowing them to fight off a future viral attack more quickly.
This new method lets us obtain information directly from the gut, without having to disturb intestinal functions. As such, the method has major advantages over endoscopies, which can be unpleasant for patients and always involves disturbing intestinal function, as the bowels need to be empty for the examination.
Professor Andrew Macpherson
To use this mechanism as a data logger, the researchers focused on something other than viral intruder DNA snippets: the mechanism can be exploited so that bacteria incorporate snippets of their own messenger RNA (mRNA) into the CRISPR array. mRNA molecules serve as the blueprint for protein production in cells. As a result, mRNA snippets can reveal which genes are being used to construct proteins that perform cellular functions.
To make the method effective, the scientists introduced the CRISPR array of the bacterial species Fusicatenibacter saccharivorans into a strain of the intestinal bacterium Escherichia coli, which is regarded as safe in humans and available as a probiotic. The transfer included the blueprint of an enzyme called reverse transcriptase, which is able to transcribe RNA into DNA. This enzyme also transcribes the information in the mRNA into DNA form, which along with accompanying CRISPR-associated proteins is necessary for incorporating the DNA snippet into the CRISPR array.
Obtaining information without disturbing the body
The modified gut bacteria were then administered to mice in the lab by researchers from the University Hospital of Bern and the University of Bern, led by Andrew Macpherson. They took feces from the animals and isolated the bacterial DNA, which they then examined using high-throughput DNA sequencing. They were able to work through the mass of data and reconstruct the genetic information of the mRNA snippets using a subsequent bioinformatic evaluation, which was performed and assessed collaboratively. This allowed the scientists to determine how frequently the gut bacteria manufactured a given mRNA molecule during their time in the body, and thus which genes are active, using noninvasive methods.
“This new method lets us obtain information directly from the gut, without having to disturb intestinal functions,” says Andrew Macpherson, Professor and Director of Gastroenterology at University Hospital Bern. As such, the method has major advantages over endoscopies, which can be unpleasant for patients and always involves disturbing intestinal function, as the bowels need to be empty for the examination.
Determining dietary status
“Bacteria are very good at registering environmental conditions and adapting their metabolism to new circumstances,” Macpherson says. The researchers were able to demonstrate how bacteria adapted their metabolism to the respective nutrient supply in experiments with mice fed different foods. The findings were reported in the most recent issue of the journal Science.
The researchers hope to improve the method so that they can one day study human patients to see how diet affects the gut ecosystem and how this affects health. They hope to use the method to determine the dietary status of children and adults in the future. Armed with this information, doctors will be able to diagnose malnutrition or decide whether a patient needs nutritional supplements.
In addition, the researchers were able to recognise inflammatory responses in the gut. The researchers administered the sensor bacteria to mice with intestinal inflammation as well as to healthy mice. In this way, they could identify the specific mRNA profile of gut bacteria that switch to inflammation mode.
Distinguishing different bacteria
The current study, published in the journal Science, includes a scientific breakthrough that allows the researchers to differentiate two strains of bacteria based on individual genetic “barcodes.” This will allow researchers to study the function of gene mutations in bacteria in laboratory animals in the future. Scientists will be able to compare the mRNA profiles of different bacteria, such as normal versus mutant bacteria, as a result of this.
For the first time, this profile can be determined as the bacteria pass through the intestine, not just when they reach the feces, so that the information shows what was happening when the bacteria were still living in the gut.
Another possible path would be to improve the system’s ability to distinguish RNA profiles of bacteria in the small and large intestines. Furthermore, the data logger function could be incorporated into other types of bacteria. This would pave the way for applications in environmental monitoring. An analysis of soil bacteria from a crop field, for example, would reveal whether herbicides were used.