Obesity is linked to a complicated inflammatory response in adipose (fat) tissue. A new study uses single-cell gene expression analysis paired with spatial transcriptomics to uncover previously unknown immune cell types and interactions within adipose tissue.
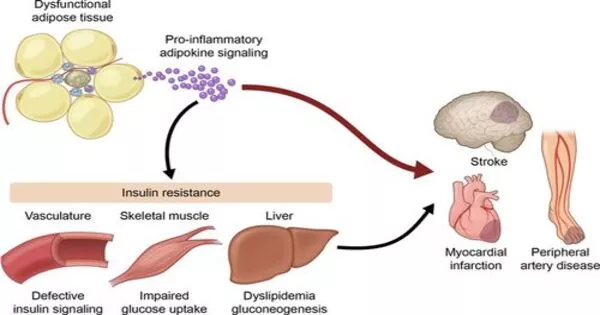
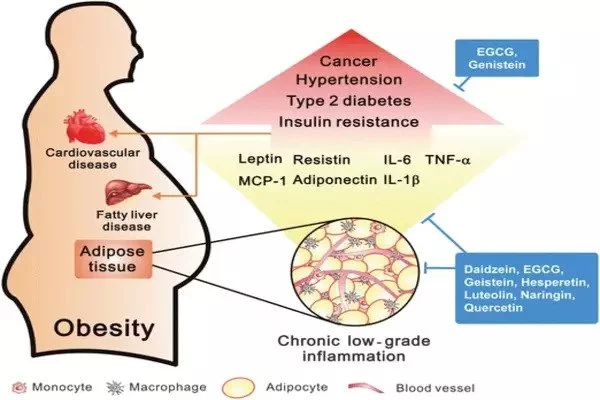
Despite its demonization, fat tissue is an extraordinarily complicated and necessary biological component involved in energy storage and hormone production, among other things. Nonetheless, modern lifestyles have resulted in a worldwide obesity epidemic, as well as an increase in linked illnesses such as type 2 diabetes and cardiovascular disease.
Researchers are aiming to explore the fundamentals of how fat tissue is organized, as well as the inflammation associated with obesity, in the hopes of unraveling the link between fat accumulation and poor health outcomes.
Lindsey Muir, Ph.D., Ph.D.-candidate Cooper Stansbury and colleagues used single-cell gene expression analysis paired with spatial transcriptomics to uncover previously unknown immune cell types and interactions within adipose tissue. Spatial transcriptomics is a relatively recent technology that detects all gene expression in small patches across a narrow piece of tissue.
Once crown-like structures appear, it takes them a long time to go away and their appearance indicates tissue dysfunction. Using image processing, we identified based on the density of these regions what was likely to be a crown-like structure and then went in to verify that we could see them visually,
Lindsey Muir
Fat research is easier said than done. In tissues that have distinct layers, such as the spinal cord or the brain, “It’s easier to do sanity checks with your data and identify this or that layer as a particular cell type and know that it should be expressing genes X, Y, and Z,” Muir, a research assistant professor in the Department of Computational Medicine and Bioinformatics, explained.
“With adipose, it’s a lot more challenging because the cell types are distributed evenly throughout the tissue, without defined cell layers.” Fat cells, or adipocytes, increase with obesity and can approach a limit, resulting in cell death and inflammation.
To better understand the types of immune cells within adipose tissue and where they are in relation to each other in obesity, the team fed mice a high-fat diet over the course of 14 weeks, collected fat tissue, and then used single-cell and spatial analyses to produce a readout of all the mRNAs present in the sample.
They were able to group cells whose genetic makeup resembled each other more closely than other groups or the overall sample by using a computational approach known as clustering in single-cell data. They observed something startling about the macrophage population in the samples, an immune cell whose function is to sweep up dead cells and debris.
“We knew going in that macrophages would likely have multiple subtypes… what surprised us were the number that came out that were highly different from each other and coming up at different times and becoming more dominant over time.”
They found five types, which they designated Mac1, 2, 3, 4, and 5. Mac1 was found in both lean and obese mice on a typical diet. Mac2 and Mac3, which were identified by their pro-inflammatory genes, peaked after 8 weeks of the high-fat diet. However, as the high-fat diet advanced to 14 weeks, Mac4 and Mac5 cells, which had low pro-inflammatory gene expression, predominated, while pro-inflammatory Mac2 and Mac3 cells declined.
“The thinking in the field has been that the type of macrophages that accumulate in obesity are promoting an inflammatory state. Based on these data there’s a lot more to the story,” said Muir. Her hypothesis is that Mac4 and Mac5 are the lipid-associated macrophages (LAMs) described in her own prior work and by other researchers and may be a sign of the body attempting to quell a damaging level of inflammation from pro-inflammatory macrophages and dying adipocytes.
Following that, painstakingly precise sectioning of fresh frozen fat tissue allowed for spatial transcriptomics investigation. In the spatial technique, each analysis site contains a unique barcode that adheres to the mRNA in the tissue on top of that spot, allowing gene expression to be mapped to precise regions in the tissue using the barcodes as coordinates. In this procedure, the sections are also photographed soon before mRNA collection. The researchers investigated these images for tell-tale indicators known as crown-like structures, which are linked to insulin resistance.
“Once crown-like structures appear, it takes them a long time to go away and their appearance indicates tissue dysfunction,” he said. “Using image processing, we identified based on the density of these regions what was likely to be a crown-like structure and then went in to verify that we could see them visually,” Muir said in a statement. Gene expression indicated the presence of Mac4 and Mac5 LAMs in these structures.