Cell surface receptors are specialized proteins located on the outer surface of the cell membrane. They play a crucial role in cell communication and signal transduction, allowing cells to respond to various external stimuli and signals from the environment.
A team of researchers from the Universities of Amsterdam and Zurich together with the Swiss company Allocyte Pharmaceuticals has for the first time been able to discover allosteric sites in a type of cell surface receptor called integrin. These receptors can detect and bind to specific molecules, such as hormones, neurotransmitters, growth factors, and other ligands.
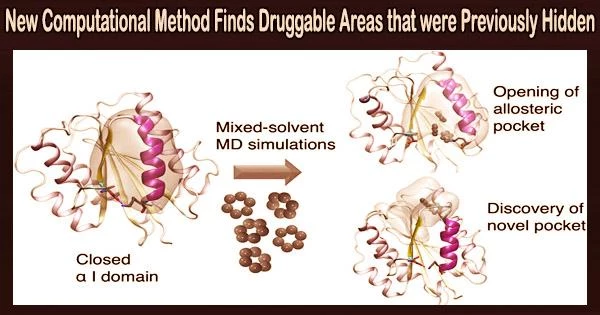
In a paper recently published in the Journal of Chemical Information and Modeling, they describe how this reveals previously inaccessible druggable integrin pockets. The research is based on a unique computational method for mixed-solvent molecular dynamics simulation created by Dr. Ioana Ilie at the University of Amsterdam’s Van ‘t Hoff Institute for Molecular Sciences’ Computational Chemistry group.
A family of cell surface adhesion receptors called integrins can send signals across membranes in both directions. They are well known for having medicinal potential for treating a variety of illnesses.
However, unanticipated downstream consequences have had an impact on the development of integrin targeting drugs. These are most frequently seen with medications that target the integrin’s native binding location.
A potential solution to these restrictions is the so-called allosteric regulation of integrins. Here, the medication attaches to the receptor in a different location, altering its conformation and affecting the protein’s ability to function.
As a result, options for drug discovery and development that may be superior to traditional orthosteric modulation are created via allosteric modulation of receptors.
Novel druggable pockets
To enable the delicate opening of the integrin α I domain, Ioana Ilie’s unique computational strategy depends on loading the solvent with tiny organic molecules (benzene in low concentrations). This revealed novel previously inaccessible druggable pockets within the integrins LFA-1, VLA-1, and Mac-1.
Thus, this study provides structural and dynamic insight into how minor changes in solvent conditions can affect new, potentially druggable pockets’ accessibility, which is then confirmed using virtual screening.
The research serves as a proof-of-concept and lays the groundwork for the development of new integrin-targeting medications. It also opens up new research directions for finding allosteric sites in other modern, undruggable protein targets.
The study also goes beyond drug discovery because it shows how even small variations in the solvent’s properties can have a significant effect on the solvated molecule’s conformational space.
This provides the chance to modify the solvent’s properties in order to get a certain response from the solvated molecule (such a protein or material), which implicitly can help create new bio-inspired materials with responsive features.