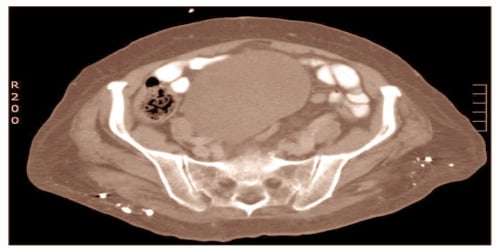
Anindya Dutta, MBBS, Ph.D., and colleagues have revealed a novel type of gene regulation that is disrupted in bladder cancer, resulting in the boosting of a gene pathway that aids the cancer cells in surviving when they are growing quickly.
Their research focuses on a 22-base transfer RNA fragment called a tRF-3b, which is altered by the enzyme complex TRMT6/61A. The methyltransferase enzyme TRMT6/61A, which adds a methyl group to the fourth base of the tRF-3bs, is found in higher concentrations in bladder cancer.
Because of this alteration, the expression of several genes involved in the unfolded protein response pathway in cancer cells is enhanced rather than silenced by tRF-3bs.
“To the best of our knowledge, this is the first example of microRNA-like gene silencing being regulated by the TRMT6/61 based on an N1-methyladenosine modification, and our report provides a mechanism by which the elevation of TRMT6/61A seen in cancers can impact gene expression,” Dutta said.
“Fast proliferating cancer cells synthesize and fold many more proteins than normal cells and thus need to upregulate the unfolded protein response pathway to maintain protein homeostasis. We find that one way bladder cancer cells activate the pro-survival unfolded protein response to alleviate endoplasmic reticulum stress is by preventing tRFs from silencing the expression of genes involved in this unfolded protein response.”
“The unfolded protein response is tightly linked to many aspects of cancer progression and has emerged as a promising therapeutic target,” Dutta said. “It has been previously noted that unfolded protein response-related genes are globally upregulated in several cancer types, including bladder cancer, and so our results suggest that inhibiting the TRMT6/61A enzyme may be a new approach to treat bladder cancer.”
The study by Dutta and co-corresponding author Rune Ougland, M.D., Ph.D., included analysis of bladder cancer tissue obtained from patients undergoing transurethral resection of bladder tumors. It is published in the journal Nature Communications.
The unfolded protein response is tightly linked to many aspects of cancer progression and has emerged as a promising therapeutic target. It has been previously noted that unfolded protein response-related genes are globally upregulated in several cancer types, including bladder cancer, and so our results suggest that inhibiting the TRMT6/61A enzyme may be a new approach to treat bladder cancer.
Anindya Dutta
Dutta is chair of the University of Alabama at Birmingham Department of Genetics, and Ougland is a urologic surgeon and senior research investigator at Oslo University Hospital Rikshospitalet, Oslo, Norway.
The process utilized to produce a library of short RNAs with the N1-methyladenosine modification, or m1A, from human cells represented a significant advancement in the study. The workflow combined two separate techniques: enrichment by m1A-antibody, small RNA sequencing, and sequencing for the m1A-induced mismatch signature.
However, the use of two additional reverse transcriptases in the workflow revealed that tRNA-derived fragments, including tRF-3b, were enriched among short RNAs. The UAB researchers discovered that a reverse transcriptase enzyme, ProtoScriptII, commonly used for short RNA sequencing, did a poor job of detecting small RNAs that contain m1A. This indicated that most small RNA-sequence libraries that frequently used ProtoScriptII were under-represented in small RNAs with a m1A mutation.
The researchers discovered that among the human short RNAs, the m1A alteration was most prevalent on tRFs thanks to the improved procedure. They also discovered that the m1A modification was very particular and frequent on both nuclear- and mitochondrial-encoded tRFs, and that it was mediated by the TRMT6/61A complex on tRF-3b from nuclear-encoded tRNAs.
How does the m1A-tRF-3b impede gene silencing?
The answer requires a further exploration of molecular genetics, although it appears that the N1-methyladenosine alteration interferes with typical Watson-Crick base pairing.
MicroRNAs are known to silence genes by binding to the RNA-induced silencing complex, or RISC. There they act as a template to bind complementary messenger RNAs, and the messenger RNA is then silenced and degraded by RISC.
In many biological pathways, including gene-silencing pathways that depend on base pairing between the short RNAs, in this case tRF-3s, and the target RNAs, tRF-3s have been discovered, similar to how microRNAs have.
Using a luciferase reporter experiment, the researchers discovered that an unmodified tRF-3 activated gene silencing while a m1A-modified tRF-3b prevented it.
“Since m1A interrupts canonical base pairing, we hypothesize the weakened base pairing by m1A in the tRF-3 with target messenger RNA explains the lowered gene-silencing activity observed for m1A-containing tRF-3s,” Dutta said.