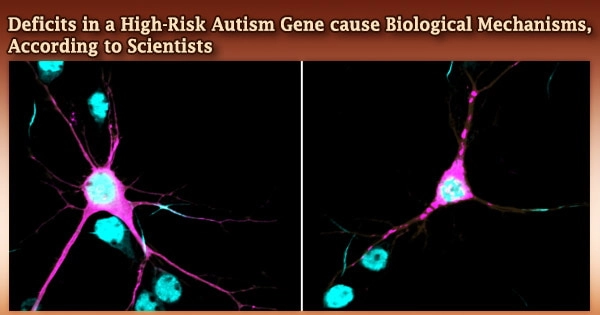
Rare variants in the ANK2 gene, which are consistently found in people with autism spectrum disorder (ASD), have been shown to alter the architecture and organization of neurons, potentially contributing to autism and neurodevelopmental comorbidities, according to researchers at the University of North Carolina at Chapel Hill School of Medicine and colleagues.
Damaris Lorenzo, PhD, an assistant professor in the UNC Department of Cell Biology and Physiology and a member of the UNC Neuroscience Center and the UNC Intellectual and Developmental Disabilities Research Center, led the study, which was published in the journal eLife.
ANK2 teaches neurons and other cell types how to create ankyrin-B, a protein that has several functions in the nervous system. Through a process known as alternative splicing, ANK2 encodes for multiple variants (isoforms) of ankyrin-B, in which sections of the protein are omitted from the final molecules.
The full-size (giant) ankyrin-B isoform is found only in neurons in mammals like mice and humans; another highly prevalent variant half its size is found in practically every type of cell and organ.
Multiple genetic investigations have consistently shown rare variations in ANK2 in people with ASD, making it one of the most reliable risk genes for the disorder. ANK2 variations can influence only gigantic ankyrin-B or both isoforms at the same time, depending on their nature and placement in the gene.
“Together with its high prevalence and striking clinical presentation, ASD’s uncertain cause is a major limiting step in advancing therapeutic options,” Lorenzo said. “The evidence of ASD’s genetic origin is strong but also complex, with at least 100 other high-risk genes linked to the disorder.”
The discovery of the origin of ASD is confounded further by the fact that single genes like ANK2 and the isoforms they encode might have several physiological activities. Scientists have identified convergent processes that may be primarily affected in people with ASD based on how subgroups of these genes overlap functionally or cooperate together to initiate biological pathways.
Neuron communication is one of these basic mechanisms, which is influenced in part by changes in axons, the long projections that transport signals from neurons to other neurons.
Together with its high prevalence and striking clinical presentation, ASD’s uncertain cause is a major limiting step in advancing therapeutic options. The evidence of ASD’s genetic origin is strong but also complex, with at least 100 other high-risk genes linked to the disorder.
Damaris Lorenzo
The axonal cytoskeleton, a complex network of filament-like proteins that plays a key role in each neuron’s growth, shape, and plasticity, is at the heart of these events within a single neuron. Another key functional axis likely to be disrupted in ASD is the axonal cytoskeleton.
The simultaneous loss of both major isoforms of ankyrin-B in the brain of mice resulted in substantial structural anomalies involving axonal wiring, highlighting the role of ankyrin-B in brain architecture and function, according to previous work by Lorenzo published in JCB.
Lorenzo and colleagues from Duke University discovered that removing only the large ankyrin-B isoform from neurons cultivated in the lab resulted in more axon branches, implying that microtubule dynamics, an important cytoskeleton component, were disrupted.
The Lorenzo group discovered that selective deletion of the big ankyrin-B isoform causes additional axon branches and volumetric increases in several axonal bundles, including the corpus callosum, in mouse brains.
They discovered that the giant ankyrin-B isoform is required to maintain the topographic order of callosal axons arising from the somatosensory cortex during brain development, as well as to ensure the specific targeting and refinement of callosal projections on the opposite side of the brain, in collaboration with Eva Anton, PhD, of the UNC Neuroscience Center and co-author of the eLife paper.
In a new mouse model that simply lacks the shorter ankyrin-B isoform, the researchers found no increases in axon branching.
“These findings confirm divergent roles of ankyrin-B isoforms and support critical and specialized roles of giant ankyrin-B in axon collateral branch formation, targeting, and refinement,” Lorenzo said.
The researchers confirmed that the observed corpus callosum abnormalities did not involve changes in myelination or the number and maturation of oligodendrocytes, a non-neuronal brain cell type implicated in similar pathologies, in collaboration with a team led by Meng Meng Fu, PhD, at the National Institute of Neurological Disorders and Stroke and co-authors of the eLife paper.
“Brain cortical regions have been the most directly linked to ASD pathology. The changes we observed in cortical structural connectivity likely result from combined defects in axon branch initiation, guidance, and pruning of misdirected or overabundant projections during development due to giant ankyrin-B deficiency,” Lorenzo said.
External cues regulate these processes, causing alterations in neurons via attractive and repulsive effects. The repulsive actions of semaphorin 3A, a protein that interacts with and collapses the terminals of axons and their branches, require the large ankyrin-B isoform in cortical neurons, according to Lorenzo’s study team.
The researchers also discovered that ANK2 variations that only affect large ankyrin-B exhibit a comparable loss of reactivity to the Semaphorin 3A protein, suggesting a probable mechanism for ASD.
“Our new insights together with our tools and methods will help us assign pathogenicity to other ANK2 variants. We are certain there is undiscovered biology relevant to brain function and ASD involving this gene and we are pursuing it,” Lorenzo said.
“Our bottom-up approach of discovery and functional validation contributes to the underdeveloped knowledge database of ASD functional etiology. This is critical because this heterogeneous and complex disorder likely requires personalized strategies for clinical intervention.”
Members of the Lorenzo lab who are co-authors in the eLife paper are co-first author Blake Creighton, lab research technician in the Lorenzo lab at the time of this research; co-first author Simone Afriyie, postbaccalaureate trainee; Deepa Ajit, PhD, a postdoctoral fellow; Kayleigh Voos, graduate student; April Burch, lab research technician; and Julia Bay, an undergraduate student at UNC-Chapel Hill. Other UNC-Chapel Hill collaborators and co-authors in the paper are Cristine Casingal, PhD, a postdoctoral fellow in the Anton research group; and Eva Anton, PhD, a research group leader.
The National Institutes of Health provided funding for this study.