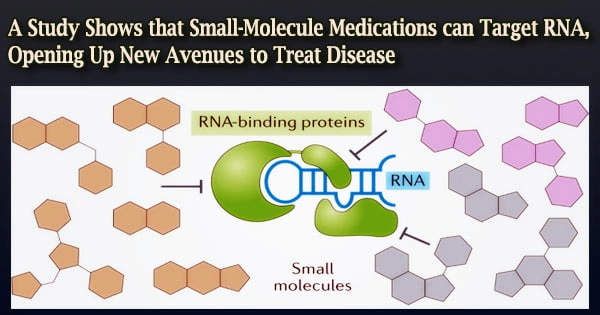
A new study published in the journal Nature provides compelling evidence that RNA (ribonucleic acid) may be a promising target for medication development. RNA plays several roles in human health. The Massachusetts General Hospital (MGH) research team’s findings shows that thousands of new biological parameters can be targeted, ushering in a new era in medication creation.
Out of the roughly 20,000 human proteins identified by the Human Genome Project, nearly all medications now on the market target one of the 700 or so proteins associated with specific diseases. However, there has been an increase in interest in recent years to include RNA to the list of “druggable” targets.
DNA (deoxyribonucleic acid) carries the genetic instructions for making proteins within cells. A section of DNA gets replicated, or “transcribed,” into a “coding” RNA, and then that RNA is converted into protein. However, 98 percent of the RNA in the human genome is “non-coding,” making up the great bulk.
“These noncoding RNAs play very important roles in the genome, and we now understand that mutations in this noncoding space can result in disease,” says the senior author of the Nature paper, Jeannie Lee, MD, PhD, of the Department of Molecular Biology at MGH.
“And there may be far more of these RNA genes than there are protein-coding genes. If we could target these RNAs, we would hugely increase the universe in which we can find drugs to treat patients.”
However, RNA hasn’t historically been a popular medication target among the pharmaceutical sector. Proteins are ideal targets because they often have stable conformations: Similar to a key in a lock, drugs latch onto proteins. Lee argues that RNA, in contrast, has a propensity to be extremely flexible, or “floppy,” and capable of taking a variety of conformations.
“If a lock is constantly changing shape, your key is not going to work,” says Lee.
Companies are reluctant to spend in trying to create drugs that target noncoding RNA due to its fragile nature. Despite all of this shape-shifting, it is known that some portions of RNA maintain stable conformations, but identifying these regions has been difficult.
These noncoding RNAs play very important roles in the genome, and we now understand that mutations in this noncoding space can result in disease. And there may be far more of these RNA genes than there are protein-coding genes. If we could target these RNAs, we would hugely increase the universe in which we can find drugs to treat patients.
Jeannie Lee
The X-chromosome inactivation (XCI), a biological mechanism that deactivates one copy of the X chromosome in female animals and is essential for normal development, is studied by Lee and her colleagues at the Massachusetts General Hospital (MGH).
Researchers from Lee’s group and Merck Research Laboratories worked together in a study directed by postdoctoral scholar Rodrigo Aguilar, PhD, to determine whether RNA would be a promising medicinal target. The Xist noncoding RNA, which silences genes on the X chromosome, was the subject of the investigation.
Finding a means to disrupt this process and awaken a dormant X chromosome could assist direct the creation of therapies for genetic conditions known as X-linked diseases, such as Rett syndrome and Fragile X syndrome, which are caused by abnormalities on the X chromosome.
The MGH researchers screened Xist against a library of 50,000 small molecule compounds with the assistance of Merck scientists Kerrie Spencer and Elliott Nickbarg, and discovered many that bind to a region on Xist known as Repeat A (RepA).
One substance, designated X1 by Lee’s team, stood out for its ability to stop multiple important proteins, including PRC2 and SPEN, from binding to RepA, which is required for Xist to silence the X chromosome.
“As a result, X inactivation cannot take place,” says Lee. To understand why, the team collaborated with structural biologists led by Trushar Patel of the University of Lethbridge in Canada.
RepA from Xist can typically take on 16 distinct conformations, whereas X1 made it take on a more consistent form. This structural alteration made it impossible for RepA to bind to PRC2 and SPEN. This study’s methodology could be utilized to locate further RNA-targeting medications.
“This really opens up a large universe for new drug development,” says Lee. “Now we don’t just have 700 proteins to target using small molecules. In the future, we may have tens and possibly hundreds of thousands of RNAs to target to cure disease.”
In addition, Lee teaches genetics at Harvard Medical School. At the moment, Aguilar works as a researcher and assistant professor at Chile’s Andres Bello University in Santiago.
The Pew Charitable Trust Latin American Fellows Program, the Howard Hughes Medical Institute, and the MGH Fund for Medical Discovery provided funding for this research.